John Dagdelen
Extracting Structured Seed-Mediated Gold Nanorod Growth Procedures from Literature with GPT-3
Apr 26, 2023Abstract:Although gold nanorods have been the subject of much research, the pathways for controlling their shape and thereby their optical properties remain largely heuristically understood. Although it is apparent that the simultaneous presence of and interaction between various reagents during synthesis control these properties, computational and experimental approaches for exploring the synthesis space can be either intractable or too time-consuming in practice. This motivates an alternative approach leveraging the wealth of synthesis information already embedded in the body of scientific literature by developing tools to extract relevant structured data in an automated, high-throughput manner. To that end, we present an approach using the powerful GPT-3 language model to extract structured multi-step seed-mediated growth procedures and outcomes for gold nanorods from unstructured scientific text. GPT-3 prompt completions are fine-tuned to predict synthesis templates in the form of JSON documents from unstructured text input with an overall accuracy of $86\%$. The performance is notable, considering the model is performing simultaneous entity recognition and relation extraction. We present a dataset of 11,644 entities extracted from 1,137 papers, resulting in 268 papers with at least one complete seed-mediated gold nanorod growth procedure and outcome for a total of 332 complete procedures.
Structured information extraction from complex scientific text with fine-tuned large language models
Dec 10, 2022



Abstract:Intelligently extracting and linking complex scientific information from unstructured text is a challenging endeavor particularly for those inexperienced with natural language processing. Here, we present a simple sequence-to-sequence approach to joint named entity recognition and relation extraction for complex hierarchical information in scientific text. The approach leverages a pre-trained large language model (LLM), GPT-3, that is fine-tuned on approximately 500 pairs of prompts (inputs) and completions (outputs). Information is extracted either from single sentences or across sentences in abstracts/passages, and the output can be returned as simple English sentences or a more structured format, such as a list of JSON objects. We demonstrate that LLMs trained in this way are capable of accurately extracting useful records of complex scientific knowledge for three representative tasks in materials chemistry: linking dopants with their host materials, cataloging metal-organic frameworks, and general chemistry/phase/morphology/application information extraction. This approach represents a simple, accessible, and highly-flexible route to obtaining large databases of structured knowledge extracted from unstructured text. An online demo is available at http://www.matscholar.com/info-extraction.
COVIDScholar: An automated COVID-19 research aggregation and analysis platform
Dec 07, 2020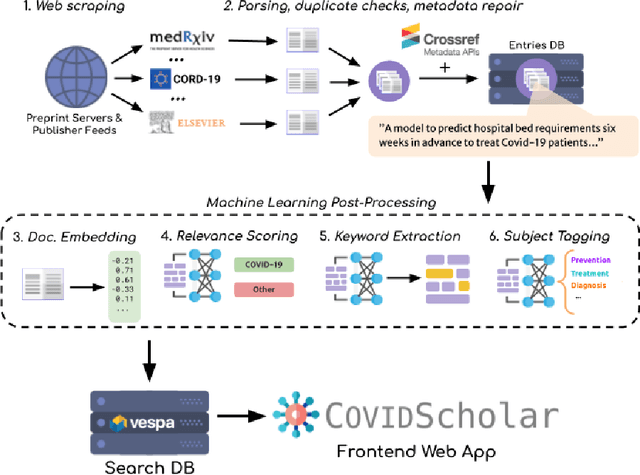
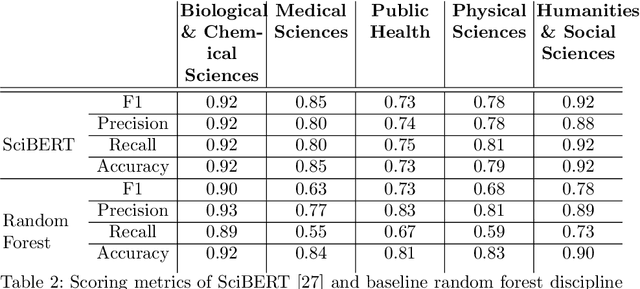
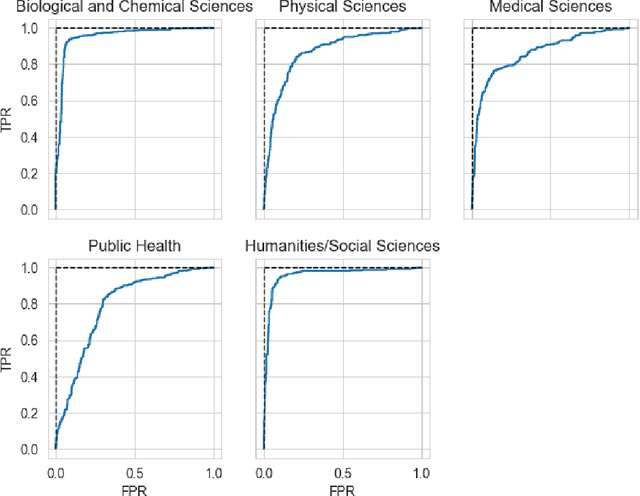
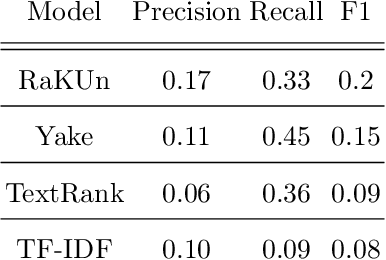
Abstract:The ongoing COVID-19 pandemic has had far-reaching effects throughout society, and science is no exception. The scale, speed, and breadth of the scientific community's COVID-19 response has lead to the emergence of new research literature on a remarkable scale -- as of October 2020, over 81,000 COVID-19 related scientific papers have been released, at a rate of over 250 per day. This has created a challenge to traditional methods of engagement with the research literature; the volume of new research is far beyond the ability of any human to read, and the urgency of response has lead to an increasingly prominent role for pre-print servers and a diffusion of relevant research across sources. These factors have created a need for new tools to change the way scientific literature is disseminated. COVIDScholar is a knowledge portal designed with the unique needs of the COVID-19 research community in mind, utilizing NLP to aid researchers in synthesizing the information spread across thousands of emergent research articles, patents, and clinical trials into actionable insights and new knowledge. The search interface for this corpus, https://covidscholar.org, now serves over 2000 unique users weekly. We present also an analysis of trends in COVID-19 research over the course of 2020.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge