Joaquin Gajardo
Wheat3DGS: In-field 3D Reconstruction, Instance Segmentation and Phenotyping of Wheat Heads with Gaussian Splatting
Apr 09, 2025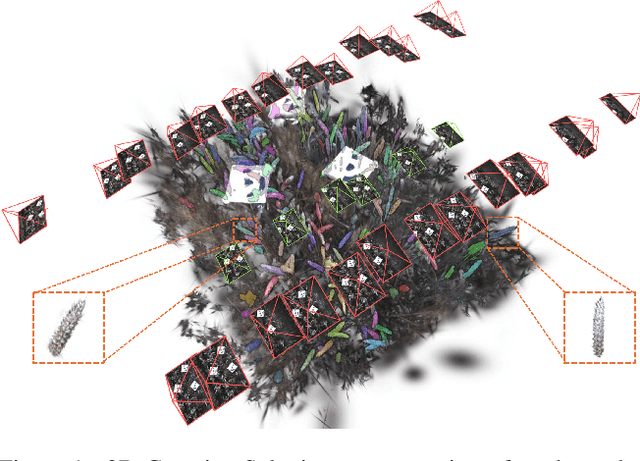
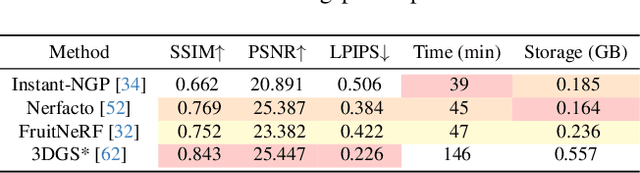
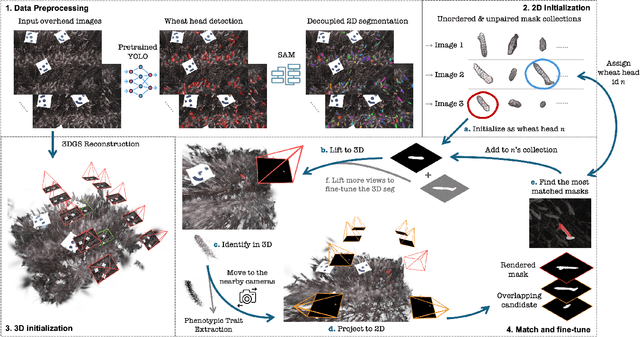

Abstract:Automated extraction of plant morphological traits is crucial for supporting crop breeding and agricultural management through high-throughput field phenotyping (HTFP). Solutions based on multi-view RGB images are attractive due to their scalability and affordability, enabling volumetric measurements that 2D approaches cannot directly capture. While advanced methods like Neural Radiance Fields (NeRFs) have shown promise, their application has been limited to counting or extracting traits from only a few plants or organs. Furthermore, accurately measuring complex structures like individual wheat heads-essential for studying crop yields-remains particularly challenging due to occlusions and the dense arrangement of crop canopies in field conditions. The recent development of 3D Gaussian Splatting (3DGS) offers a promising alternative for HTFP due to its high-quality reconstructions and explicit point-based representation. In this paper, we present Wheat3DGS, a novel approach that leverages 3DGS and the Segment Anything Model (SAM) for precise 3D instance segmentation and morphological measurement of hundreds of wheat heads automatically, representing the first application of 3DGS to HTFP. We validate the accuracy of wheat head extraction against high-resolution laser scan data, obtaining per-instance mean absolute percentage errors of 15.1%, 18.3%, and 40.2% for length, width, and volume. We provide additional comparisons to NeRF-based approaches and traditional Muti-View Stereo (MVS), demonstrating superior results. Our approach enables rapid, non-destructive measurements of key yield-related traits at scale, with significant implications for accelerating crop breeding and improving our understanding of wheat development.
Country-Scale Cropland Mapping in Data-Scarce Settings Using Deep Learning: A Case Study of Nigeria
Dec 18, 2023Abstract:Cropland maps are a core and critical component of remote-sensing-based agricultural monitoring, providing dense and up-to-date information about agricultural development. Machine learning is an effective tool for large-scale agricultural mapping, but relies on geo-referenced ground-truth data for model training and testing, which can be scarce or time-consuming to obtain. In this study, we explore the usefulness of combining a global cropland dataset and a hand-labeled dataset to train machine learning models for generating a new cropland map for Nigeria in 2020 at 10 m resolution. We provide the models with pixel-wise time series input data from remote sensing sources such as Sentinel-1 and 2, ERA5 climate data, and DEM data, in addition to binary labels indicating cropland presence. We manually labeled 1827 evenly distributed pixels across Nigeria, splitting them into 50\% training, 25\% validation, and 25\% test sets used to fit the models and test our output map. We evaluate and compare the performance of single- and multi-headed Long Short-Term Memory (LSTM) neural network classifiers, a Random Forest classifier, and three existing 10 m resolution global land cover maps (Google's Dynamic World, ESRI's Land Cover, and ESA's WorldCover) on our proposed test set. Given the regional variations in cropland appearance, we additionally experimented with excluding or sub-setting the global crowd-sourced Geowiki cropland dataset, to empirically assess the trade-off between data quantity and data quality in terms of the similarity to the target data distribution of Nigeria. We find that the existing WorldCover map performs the best with an F1-score of 0.825 and accuracy of 0.870 on the test set, followed by a single-headed LSTM model trained with our hand-labeled training samples and the Geowiki data points in Nigeria, with a F1-score of 0.814 and accuracy of 0.842.
Using Machine Learning to generate an open-access cropland map from satellite images time series in the Indian Himalayan Region
Mar 28, 2022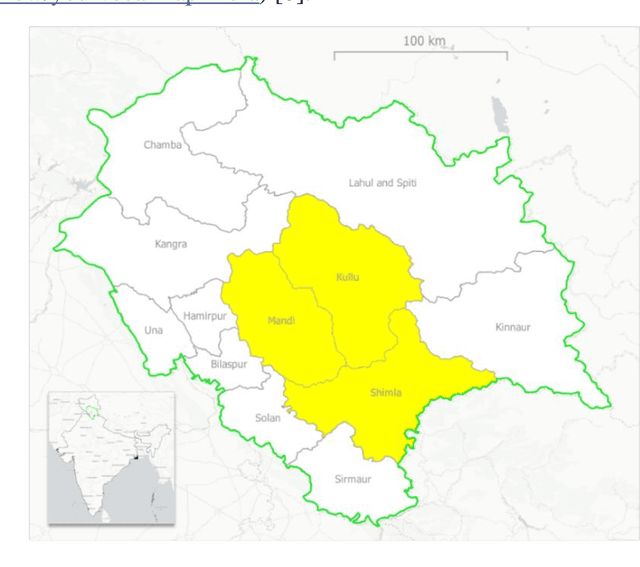
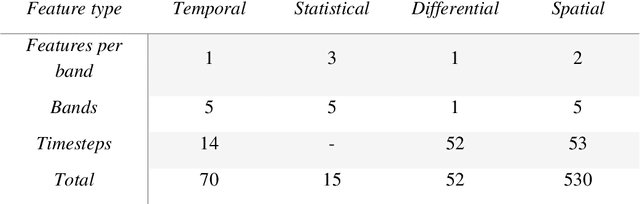
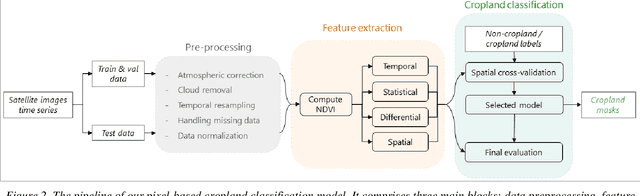
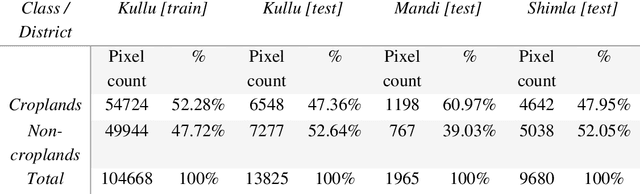
Abstract:Crop maps are crucial for agricultural monitoring and food management and can additionally support domain-specific applications, such as setting cold supply chain infrastructure in developing countries. Machine learning (ML) models, combined with freely-available satellite imagery, can be used to produce cost-effective and high spatial-resolution crop maps. However, accessing ground truth data for supervised learning is especially challenging in developing countries due to factors such as smallholding and fragmented geography, which often results in a lack of crop type maps or even reliable cropland maps. Our area of interest for this study lies in Himachal Pradesh, India, where we aim at producing an open-access binary cropland map at 10-meter resolution for the Kullu, Shimla, and Mandi districts. To this end, we developed an ML pipeline that relies on Sentinel-2 satellite images time series. We investigated two pixel-based supervised classifiers, support vector machines (SVM) and random forest (RF), which are used to classify per-pixel time series for binary cropland mapping. The ground truth data used for training, validation and testing was manually annotated from a combination of field survey reference points and visual interpretation of very high resolution (VHR) imagery. We trained and validated the models via spatial cross-validation to account for local spatial autocorrelation and selected the RF model due to overall robustness and lower computational cost. We tested the generalization capability of the chosen model at the pixel level by computing the accuracy, recall, precision, and F1-score on hold-out test sets of each district, achieving an average accuracy for the RF (our best model) of 87%. We used this model to generate a cropland map for three districts of Himachal Pradesh, spanning 14,600 km2, which improves the resolution and quality of existing public maps.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge