Jingfeng Jiang
SFD-Mamba2Net: Strcture-Guided Frequency-Enhanced Dual-Stream Mamba2 Network for Coronary Artery Segmentation
Sep 10, 2025



Abstract:Background: Coronary Artery Disease (CAD) is one of the leading causes of death worldwide. Invasive Coronary Angiography (ICA), regarded as the gold standard for CAD diagnosis, necessitates precise vessel segmentation and stenosis detection. However, ICA images are typically characterized by low contrast, high noise levels, and complex, fine-grained vascular structures, which pose significant challenges to the clinical adoption of existing segmentation and detection methods. Objective: This study aims to improve the accuracy of coronary artery segmentation and stenosis detection in ICA images by integrating multi-scale structural priors, state-space-based long-range dependency modeling, and frequency-domain detail enhancement strategies. Methods: We propose SFD-Mamba2Net, an end-to-end framework tailored for ICA-based vascular segmentation and stenosis detection. In the encoder, a Curvature-Aware Structural Enhancement (CASE) module is embedded to leverage multi-scale responses for highlighting slender tubular vascular structures, suppressing background interference, and directing attention toward vascular regions. In the decoder, we introduce a Progressive High-Frequency Perception (PHFP) module that employs multi-level wavelet decomposition to progressively refine high-frequency details while integrating low-frequency global structures. Results and Conclusions: SFD-Mamba2Net consistently outperformed state-of-the-art methods across eight segmentation metrics, and achieved the highest true positive rate and positive predictive value in stenosis detection.
FAD-Net: Frequency-Domain Attention-Guided Diffusion Network for Coronary Artery Segmentation using Invasive Coronary Angiography
Jun 13, 2025Abstract:Background: Coronary artery disease (CAD) remains one of the leading causes of mortality worldwide. Precise segmentation of coronary arteries from invasive coronary angiography (ICA) is critical for effective clinical decision-making. Objective: This study aims to propose a novel deep learning model based on frequency-domain analysis to enhance the accuracy of coronary artery segmentation and stenosis detection in ICA, thereby offering robust support for the stenosis detection and treatment of CAD. Methods: We propose the Frequency-Domain Attention-Guided Diffusion Network (FAD-Net), which integrates a frequency-domain-based attention mechanism and a cascading diffusion strategy to fully exploit frequency-domain information for improved segmentation accuracy. Specifically, FAD-Net employs a Multi-Level Self-Attention (MLSA) mechanism in the frequency domain, computing the similarity between queries and keys across high- and low-frequency components in ICAs. Furthermore, a Low-Frequency Diffusion Module (LFDM) is incorporated to decompose ICAs into low- and high-frequency components via multi-level wavelet transformation. Subsequently, it refines fine-grained arterial branches and edges by reintegrating high-frequency details via inverse fusion, enabling continuous enhancement of anatomical precision. Results and Conclusions: Extensive experiments demonstrate that FAD-Net achieves a mean Dice coefficient of 0.8717 in coronary artery segmentation, outperforming existing state-of-the-art methods. In addition, it attains a true positive rate of 0.6140 and a positive predictive value of 0.6398 in stenosis detection, underscoring its clinical applicability. These findings suggest that FAD-Net holds significant potential to assist in the accurate diagnosis and treatment planning of CAD.
AGMN: Association Graph-based Graph Matching Network for Coronary Artery Semantic Labeling on Invasive Coronary Angiograms
Jan 11, 2023



Abstract:Semantic labeling of coronary arterial segments in invasive coronary angiography (ICA) is important for automated assessment and report generation of coronary artery stenosis in the computer-aided diagnosis of coronary artery disease (CAD). Inspired by the training procedure of interventional cardiologists for interpreting the structure of coronary arteries, we propose an association graph-based graph matching network (AGMN) for coronary arterial semantic labeling. We first extract the vascular tree from invasive coronary angiography (ICA) and convert it into multiple individual graphs. Then, an association graph is constructed from two individual graphs where each vertex represents the relationship between two arterial segments. Using the association graph, the AGMN extracts the vertex features by the embedding module, aggregates the features from adjacent vertices and edges by graph convolution network, and decodes the features to generate the semantic mappings between arteries. By learning the mapping of arterial branches between two individual graphs, the unlabeled arterial segments are classified by the labeled segments to achieve semantic labeling. A dataset containing 263 ICAs was employed to train and validate the proposed model, and a five-fold cross-validation scheme was performed. Our AGMN model achieved an average accuracy of 0.8264, an average precision of 0.8276, an average recall of 0.8264, and an average F1-score of 0.8262, which significantly outperformed existing coronary artery semantic labeling methods. In conclusion, we have developed and validated a new algorithm with high accuracy, interpretability, and robustness for coronary artery semantic labeling on ICAs.
Automatic Identification of the End-Diastolic and End-Systolic Cardiac Frames from Invasive Coronary Angiography Videos
Oct 06, 2021

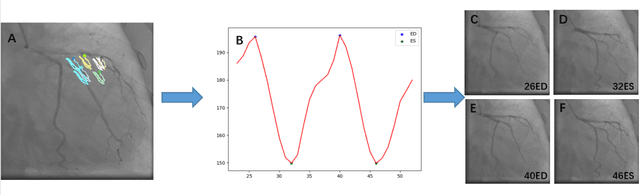
Abstract:Automatic identification of proper image frames at the end-diastolic (ED) and end-systolic (ES) frames during the review of invasive coronary angiograms (ICA) is important to assess blood flow during a cardiac cycle, reconstruct the 3D arterial anatomy from bi-planar views, and generate the complementary fusion map with myocardial images. The current identification method primarily relies on visual interpretation, making it not only time-consuming but also less reproducible. In this paper, we propose a new method to automatically identify angiographic image frames associated with the ED and ES cardiac phases by using the trajectories of key vessel points (i.e. landmarks). More specifically, a detection algorithm is first used to detect the key points of coronary arteries, and then an optical flow method is employed to track the trajectories of the selected key points. The ED and ES frames are identified based on all these trajectories. Our method was tested with 62 ICA videos from two separate medical centers (22 and 9 patients in sites 1 and 2, respectively). Comparing consensus interpretations by two human expert readers, excellent agreement was achieved by the proposed algorithm: the agreement rates within a one-frame range were 92.99% and 92.73% for the automatic identification of the ED and ES image frames, respectively. In conclusion, the proposed automated method showed great potential for being an integral part of automated ICA image analysis.
A Deep Learning-based Method to Extract Lumen and Media-Adventitia in Intravascular Ultrasound Images
Feb 21, 2021



Abstract:Intravascular ultrasound (IVUS) imaging allows direct visualization of the coronary vessel wall and is suitable for the assessment of atherosclerosis and the degree of stenosis. Accurate segmentation and measurements of lumen and median-adventitia (MA) from IVUS are essential for such a successful clinical evaluation. However, current segmentation relies on manual operations, which is time-consuming and user-dependent. In this paper, we aim to develop a deep learning-based method using an encoder-decoder deep architecture to automatically extract both lumen and MA border. Our method named IVUS-U-Net++ is an extension of the well-known U-Net++ model. More specifically, a feature pyramid network was added to the U-Net++ model, enabling the utilization of feature maps at different scales. As a result, the accuracy of the probability map and subsequent segmentation have been improved We collected 1746 IVUS images from 18 patients in this study. The whole dataset was split into a training dataset (1572 images) for the 10-fold cross-validation and a test dataset (174 images) for evaluating the performance of models. Our IVUS-U-Net++ segmentation model achieved a Jaccard measure (JM) of 0.9412, a Hausdorff distance (HD) of 0.0639 mm for the lumen border, and a JM of 0.9509, an HD of 0.0867 mm for the MA border, respectively. Moreover, the Pearson correlation and Bland-Altman analyses were performed to evaluate the correlations of 12 clinical parameters measured from our segmentation results and the ground truth, and automatic measurements agreed well with those from the ground truth (all Ps<0.01). In conclusion, our preliminary results demonstrate that the proposed IVUS-U-Net++ model has great promise for clinical use.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge