Jianbing Zhu
Efficient Training of 3D Seismic Image Fault Segmentation Network under Sparse Labels by Weakening Anomaly Annotation
Oct 19, 2021



Abstract:Seismic data fault detection has recently been regarded as a 3D image segmentation task. The nature of fault structures in seismic image makes it difficult to manually label faults. Manual labeling often has many false negative labels (abnormal annotations), which will seriously jeopardize the training process. In this work, we find that region-based loss significantly outperforms distribution-based loss when dealing with false negative labels, therefore we proposed Mask Dice loss (MD loss), which is the first reported region-based loss function for training 3D image segmentation networks using sparse 2D slice labels. In addition, fault is an edge feature, and the current network widely used for fault segmentation downsamples the features multiple times, which is not conducive to edge representation and thus requires many parameters and computational effort to preserve the features. We proposed Fault-Net, which uses a high-resolution and shallow structure to propagate multi-scale features in parallel, fully preserving edge features. Meanwhile, in order to efficiently fuse multiscale features, we decouple the convolution process into feature selection and channel fusion, and proposed a lightweight feature fusion block, Multi-Scale Compression Fusion (MCF). Because the Fault-Net always keeps the edge features during propagation, only few parameters and computation are required. Experimental results show that MD loss can clearly weaken the effect of false negative labels. The Fault-Net parameter is only 0.42MB, support up to 528^3(1.5x10^8, Float32) size cuboid inference on 16GB video ram, its inference speed on CPU and GPU is significantly faster than other networks. It works well on most of the open data seismic images, and the result of our approach is state-ofthe-art in FORCE fault identification competition.
Seismic Fault Segmentation via 3D-CNN Training by a Few 2D Slices Labels
May 20, 2021



Abstract:Detection faults in seismic data is a crucial step for seismic structural interpretation, reservoir characterization and well placement. Some recent works regard it as an image segmentation task. The task of image segmentation requires huge labels, especially 3D seismic data, which has a complex structure and lots of noise. Therefore, its annotation requires expert experience and a huge workload. In this study, we present {\lambda}-BCE and {\lambda}-smooth L1loss to effectively train 3D-CNN by some slices from 3D seismic data, so that the model can learn the segmentation of 3D seismic data from a few 2D slices. In order to fully extract information from limited data and suppress seismic noise, we propose an attention module that can be used for active supervision training and embedded in the network. The attention heatmap target is generated by the original label, and letting it supervise the attention module using the {\lambda}-smooth L1loss. The experiment proves the effectiveness of our loss function and attention module, it also shows that our method can extract 3D seismic features from a few 2D slices labels, and the segmentation effect achieves state-of-the-art. We only use 3.3% of the all labels, and we can achieve similar performance as using all labels. This work has been submitted to the IEEE for possible publication. Copyright may be transferred without notice, after which this version may no longer be accessible.
Crossover-Net: Leveraging the Vertical-Horizontal Crossover Relation for Robust Segmentation
Apr 03, 2020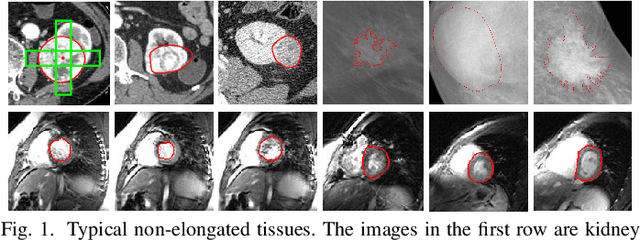
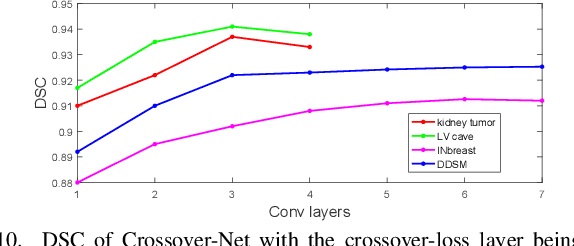
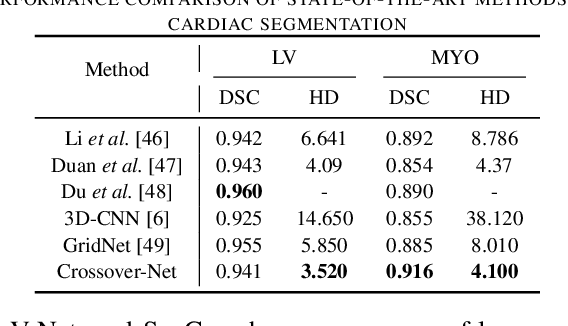
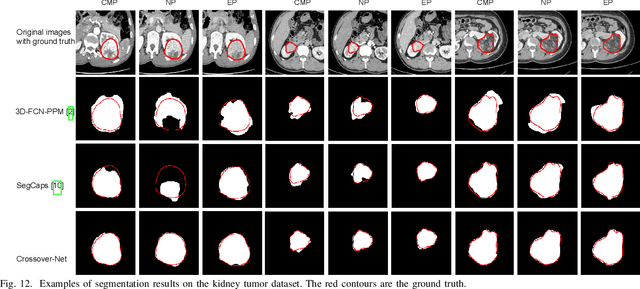
Abstract:Robust segmentation for non-elongated tissues in medical images is hard to realize due to the large variation of the shape, size, and appearance of these tissues in different patients. In this paper, we present an end-to-end trainable deep segmentation model termed Crossover-Net for robust segmentation in medical images. Our proposed model is inspired by an insightful observation: during segmentation, the representation from the horizontal and vertical directions can provide different local appearance and orthogonality context information, which helps enhance the discrimination between different tissues by simultaneously learning from these two directions. Specifically, by converting the segmentation task to a pixel/voxel-wise prediction problem, firstly, we originally propose a cross-shaped patch, namely crossover-patch, which consists of a pair of (orthogonal and overlapped) vertical and horizontal patches, to capture the orthogonal vertical and horizontal relation. Then, we develop the Crossover-Net to learn the vertical-horizontal crossover relation captured by our crossover-patches. To achieve this goal, for learning the representation on a typical crossover-patch, we design a novel loss function to (1) impose the consistency on the overlap region of the vertical and horizontal patches and (2) preserve the diversity on their non-overlap regions. We have extensively evaluated our method on CT kidney tumor, MR cardiac, and X-ray breast mass segmentation tasks. Promising results are achieved according to our extensive evaluation and comparison with the state-of-the-art segmentation models.
Crossbar-Net: A Novel Convolutional Network for Kidney Tumor Segmentation in CT Images
Aug 02, 2018



Abstract:Due to the irregular motion, similar appearance and diverse shape, accurate segmentation of kidney tumor in CT images is a difficult and challenging task. To this end, we present a novel automatic segmentation method, termed as Crossbar-Net, with the goal of accurate segmenting the kidney tumors. Firstly, considering that the traditional learning-based segmentation methods normally employ either whole images or squared patches as the training samples, we innovatively sample the orthogonal non-squared patches (namely crossbar patches), to fully cover the whole kidney tumors in either horizontal or vertical directions. These sampled crossbar patches could not only represent the detailed local information of kidney tumor as the traditional patches, but also describe the global appearance from either horizontal or vertical direction using contextual information. Secondly, with the obtained crossbar patches, we trained a convolutional neural network with two sub-models (i.e., horizontal sub-model and vertical sub-model) in a cascaded manner, to integrate the segmentation results from two directions (i.e., horizontal and vertical). This cascaded training strategy could effectively guarantee the consistency between sub-models, by feeding each other with the most difficult samples, for a better segmentation. In the experiment, we evaluate our method on a real CT kidney tumor dataset, collected from 94 different patients including 3,500 images. Compared with the state-of-the-art segmentation methods, the results demonstrate the superior results of our method on dice ratio score, true positive fraction, centroid distance and Hausdorff distance. Moreover, we have extended our crossbar-net to a different task: cardiac segmentation, showing the promising results for the better generalization.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge