Ji Hwan Park
idMotif: An Interactive Motif Identification in Protein Sequences
Feb 04, 2024



Abstract:This article introduces idMotif, a visual analytics framework designed to aid domain experts in the identification of motifs within protein sequences. Motifs, short sequences of amino acids, are critical for understanding the distinct functions of proteins. Identifying these motifs is pivotal for predicting diseases or infections. idMotif employs a deep learning-based method for the categorization of protein sequences, enabling the discovery of potential motif candidates within protein groups through local explanations of deep learning model decisions. It offers multiple interactive views for the analysis of protein clusters or groups and their sequences. A case study, complemented by expert feedback, illustrates idMotif's utility in facilitating the analysis and identification of protein sequences and motifs.
* IEEE CGA
A Bayesian Deep Learning Approach to Near-Term Climate Prediction
Feb 23, 2022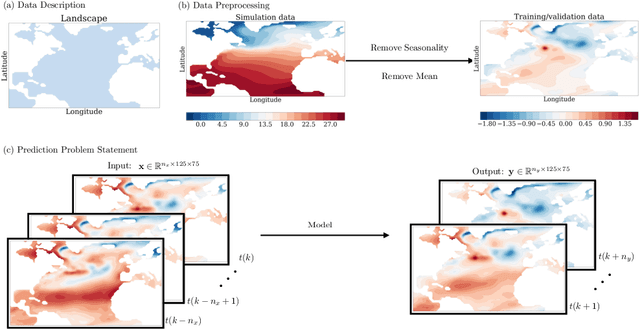
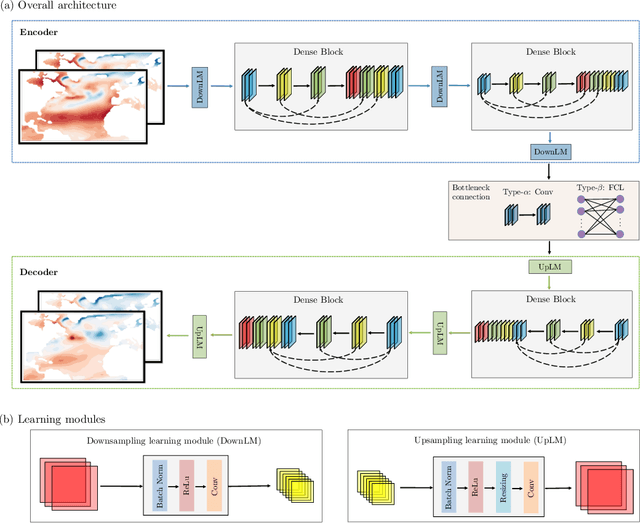
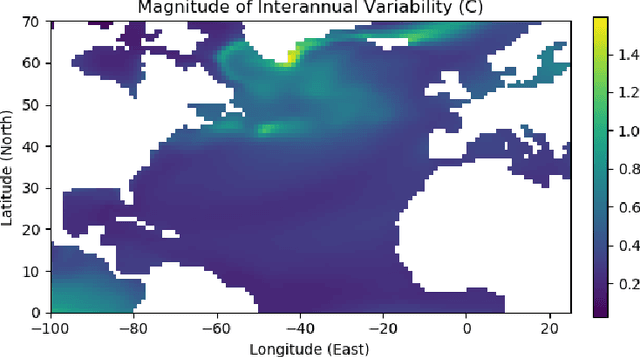
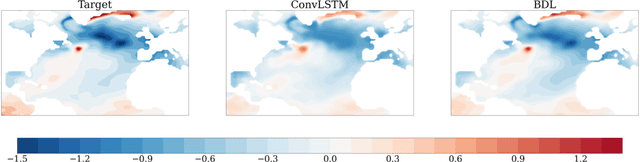
Abstract:Since model bias and associated initialization shock are serious shortcomings that reduce prediction skills in state-of-the-art decadal climate prediction efforts, we pursue a complementary machine-learning-based approach to climate prediction. The example problem setting we consider consists of predicting natural variability of the North Atlantic sea surface temperature on the interannual timescale in the pre-industrial control simulation of the Community Earth System Model (CESM2). While previous works have considered the use of recurrent networks such as convolutional LSTMs and reservoir computing networks in this and other similar problem settings, we currently focus on the use of feedforward convolutional networks. In particular, we find that a feedforward convolutional network with a Densenet architecture is able to outperform a convolutional LSTM in terms of predictive skill. Next, we go on to consider a probabilistic formulation of the same network based on Stein variational gradient descent and find that in addition to providing useful measures of predictive uncertainty, the probabilistic (Bayesian) version improves on its deterministic counterpart in terms of predictive skill. Finally, we characterize the reliability of the ensemble of ML models obtained in the probabilistic setting by using analysis tools developed in the context of ensemble numerical weather prediction.
Feature Importance in a Deep Learning Climate Emulator
Aug 27, 2021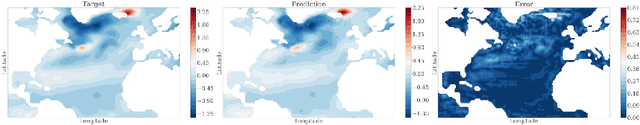
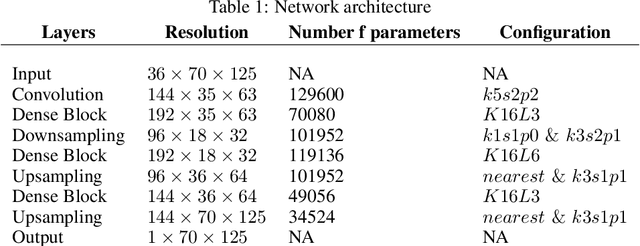
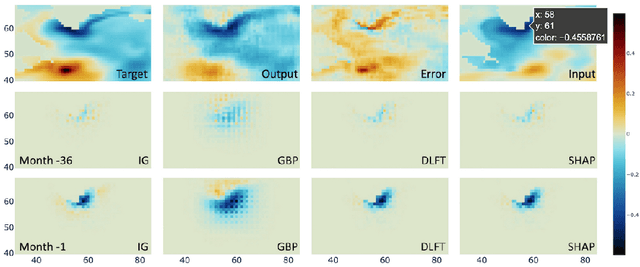
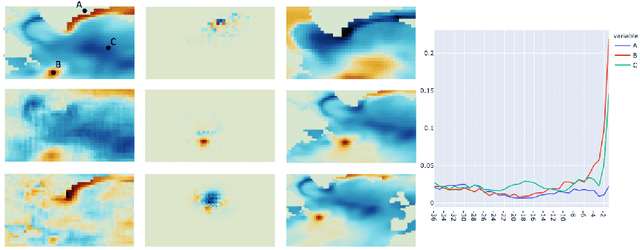
Abstract:We present a study using a class of post-hoc local explanation methods i.e., feature importance methods for "understanding" a deep learning (DL) emulator of climate. Specifically, we consider a multiple-input-single-output emulator that uses a DenseNet encoder-decoder architecture and is trained to predict interannual variations of sea surface temperature (SST) at 1, 6, and 9 month lead times using the preceding 36 months of (appropriately filtered) SST data. First, feature importance methods are employed for individual predictions to spatio-temporally identify input features that are important for model prediction at chosen geographical regions and chosen prediction lead times. In a second step, we also examine the behavior of feature importance in a generalized sense by considering an aggregation of the importance heatmaps over training samples. We find that: 1) the climate emulator's prediction at any geographical location depends dominantly on a small neighborhood around it; 2) the longer the prediction lead time, the further back the "importance" extends; and 3) to leading order, the temporal decay of "importance" is independent of geographical location. An ablation experiment is adopted to verify the findings. From the perspective of climate dynamics, these findings suggest a dominant role for local processes and a negligible role for remote teleconnections at the spatial and temporal scales we consider. From the perspective of network architecture, the spatio-temporal relations between the inputs and outputs we find suggest potential model refinements. We discuss further extensions of our methods, some of which we are considering in ongoing work.
C2A: Crowd Consensus Analytics for Virtual Colonoscopy
Oct 21, 2018



Abstract:We present a medical crowdsourcing visual analytics platform called C{$^2$}A to visualize, classify and filter crowdsourced clinical data. More specifically, C$^2$A is used to build consensus on a clinical diagnosis by visualizing crowd responses and filtering out anomalous activity. Crowdsourcing medical applications have recently shown promise where the non-expert users (the crowd) were able to achieve accuracy similar to the medical experts. This has the potential to reduce interpretation/reading time and possibly improve accuracy by building a consensus on the findings beforehand and letting the medical experts make the final diagnosis. In this paper, we focus on a virtual colonoscopy (VC) application with the clinical technicians as our target users, and the radiologists acting as consultants and classifying segments as benign or malignant. In particular, C$^2$A is used to analyze and explore crowd responses on video segments, created from fly-throughs in the virtual colon. C$^2$A provides several interactive visualization components to build crowd consensus on video segments, to detect anomalies in the crowd data and in the VC video segments, and finally, to improve the non-expert user's work quality and performance by A/B testing for the optimal crowdsourcing platform and application-specific parameters. Case studies and domain experts feedback demonstrate the effectiveness of our framework in improving workers' output quality, the potential to reduce the radiologists' interpretation time, and hence, the potential to improve the traditional clinical workflow by marking the majority of the video segments as benign based on the crowd consensus.
* IEEE Conference on Visual Analytics Science and Technology (VAST), pp. 21-30, 2016 (10 pages, 11 figures)
Crowd-Assisted Polyp Annotation of Virtual Colonoscopy Videos
Sep 17, 2018Abstract:Virtual colonoscopy (VC) allows a radiologist to navigate through a 3D colon model reconstructed from a computed tomography scan of the abdomen, looking for polyps, the precursors of colon cancer. Polyps are seen as protrusions on the colon wall and haustral folds, visible in the VC fly-through videos. A complete review of the colon surface requires full navigation from the rectum to the cecum in antegrade and retrograde directions, which is a tedious task that takes an average of 30 minutes. Crowdsourcing is a technique for non-expert users to perform certain tasks, such as image or video annotation. In this work, we use crowdsourcing for the examination of complete VC fly-through videos for polyp annotation by non-experts. The motivation for this is to potentially help the radiologist reach a diagnosis in a shorter period of time, and provide a stronger confirmation of the eventual diagnosis. The crowdsourcing interface includes an interactive tool for the crowd to annotate suspected polyps in the video with an enclosing box. Using our workflow, we achieve an overall polyps-per-patient sensitivity of 87.88% (95.65% for polyps $\geq$5mm and 70% for polyps $<$5mm). We also demonstrate the efficacy and effectiveness of a non-expert user in detecting and annotating polyps and discuss their possibility in aiding radiologists in VC examinations.
Crowdsourcing Lung Nodules Detection and Annotation
Sep 17, 2018Abstract:We present crowdsourcing as an additional modality to aid radiologists in the diagnosis of lung cancer from clinical chest computed tomography (CT) scans. More specifically, a complete workflow is introduced which can help maximize the sensitivity of lung nodule detection by utilizing the collective intelligence of the crowd. We combine the concept of overlapping thin-slab maximum intensity projections (TS-MIPs) and cine viewing to render short videos that can be outsourced as an annotation task to the crowd. These videos are generated by linearly interpolating overlapping TS-MIPs of CT slices through the depth of each quadrant of a patient's lung. The resultant videos are outsourced to an online community of non-expert users who, after a brief tutorial, annotate suspected nodules in these video segments. Using our crowdsourcing workflow, we achieved a lung nodule detection sensitivity of over 90% for 20 patient CT datasets (containing 178 lung nodules with sizes between 1-30mm), and only 47 false positives from a total of 1021 annotations on nodules of all sizes (96% sensitivity for nodules$>$4mm). These results show that crowdsourcing can be a robust and scalable modality to aid radiologists in screening for lung cancer, directly or in combination with computer-aided detection (CAD) algorithms. For CAD algorithms, the presented workflow can provide highly accurate training data to overcome the high false-positive rate (per scan) problem. We also provide, for the first time, analysis on nodule size and position which can help improve CAD algorithms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge