Jeng-Lin Li
Dual-Process Scaffold Reasoning for Enhancing LLM Code Debugging
Nov 11, 2025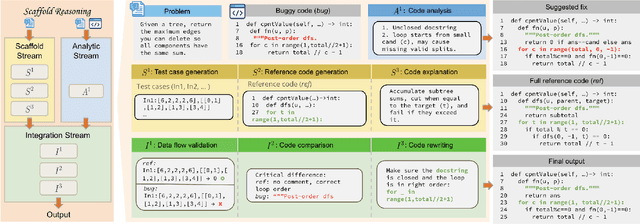
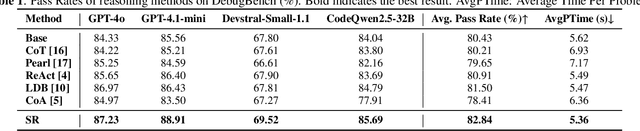
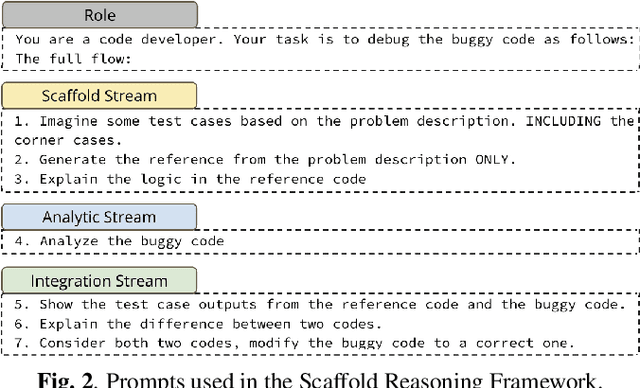
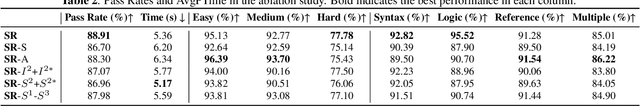
Abstract:Recent LLMs have demonstrated sophisticated problem-solving capabilities on various benchmarks through advanced reasoning algorithms. However, the key research question of identifying reasoning steps that balance complexity and computational efficiency remains unsolved. Recent research has increasingly drawn upon psychological theories to explore strategies for optimizing cognitive pathways. The LLM's final outputs and intermediate steps are regarded as System 1 and System 2, respectively. However, an in-depth exploration of the System 2 reasoning is still lacking. Therefore, we propose a novel psychologically backed Scaffold Reasoning framework for code debugging, which encompasses the Scaffold Stream, Analytic Stream, and Integration Stream. The construction of reference code within the Scaffold Stream is integrated with the buggy code analysis results produced by the Analytic Stream through the Integration Stream. Our framework achieves an 88.91% pass rate and an average inference time of 5.36 seconds per-problem on DebugBench, outperforming other reasoning approaches across various LLMs in both reasoning accuracy and efficiency. Further analyses elucidate the advantages and limitations of various cognitive pathways across varying problem difficulties and bug types. Our findings also corroborate the alignment of the proposed Scaffold Reasoning framework with human cognitive processes.
How Bias Binds: Measuring Hidden Associations for Bias Control in Text-to-Image Compositions
Nov 10, 2025Abstract:Text-to-image generative models often exhibit bias related to sensitive attributes. However, current research tends to focus narrowly on single-object prompts with limited contextual diversity. In reality, each object or attribute within a prompt can contribute to bias. For example, the prompt "an assistant wearing a pink hat" may reflect female-inclined biases associated with a pink hat. The neglected joint effects of the semantic binding in the prompts cause significant failures in current debiasing approaches. This work initiates a preliminary investigation on how bias manifests under semantic binding, where contextual associations between objects and attributes influence generative outcomes. We demonstrate that the underlying bias distribution can be amplified based on these associations. Therefore, we introduce a bias adherence score that quantifies how specific object-attribute bindings activate bias. To delve deeper, we develop a training-free context-bias control framework to explore how token decoupling can facilitate the debiasing of semantic bindings. This framework achieves over 10% debiasing improvement in compositional generation tasks. Our analysis of bias scores across various attribute-object bindings and token decorrelation highlights a fundamental challenge: reducing bias without disrupting essential semantic relationships. These findings expose critical limitations in current debiasing approaches when applied to semantically bound contexts, underscoring the need to reassess prevailing bias mitigation strategies.
Bloodroot: When Watermarking Turns Poisonous For Stealthy Backdoor
Oct 09, 2025Abstract:Backdoor data poisoning is a crucial technique for ownership protection and defending against malicious attacks. Embedding hidden triggers in training data can manipulate model outputs, enabling provenance verification, and deterring unauthorized use. However, current audio backdoor methods are suboptimal, as poisoned audio often exhibits degraded perceptual quality, which is noticeable to human listeners. This work explores the intrinsic stealthiness and effectiveness of audio watermarking in achieving successful poisoning. We propose a novel Watermark-as-Trigger concept, integrated into the Bloodroot backdoor framework via adversarial LoRA fine-tuning, which enhances perceptual quality while achieving a much higher trigger success rate and clean-sample accuracy. Experiments on speech recognition (SR) and speaker identification (SID) datasets show that watermark-based poisoning remains effective under acoustic filtering and model pruning. The proposed Bloodroot backdoor framework not only secures data-to-model ownership, but also well reveals the risk of adversarial misuse.
Sharpness-Aware Geometric Defense for Robust Out-Of-Distribution Detection
Aug 24, 2025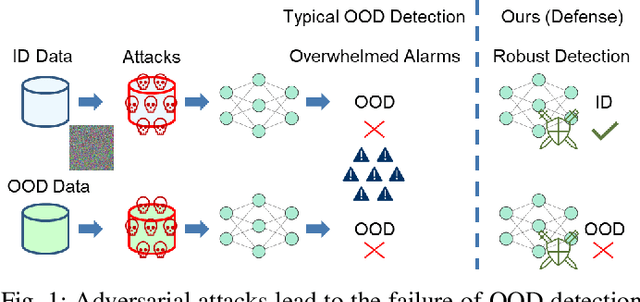
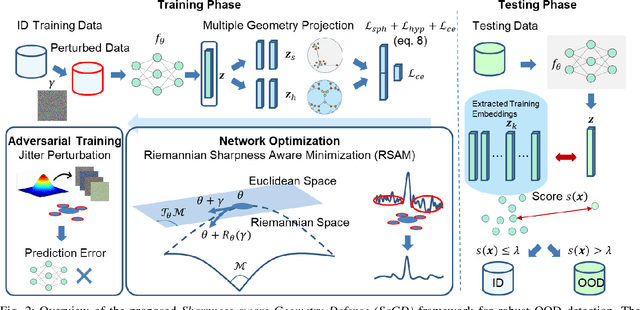
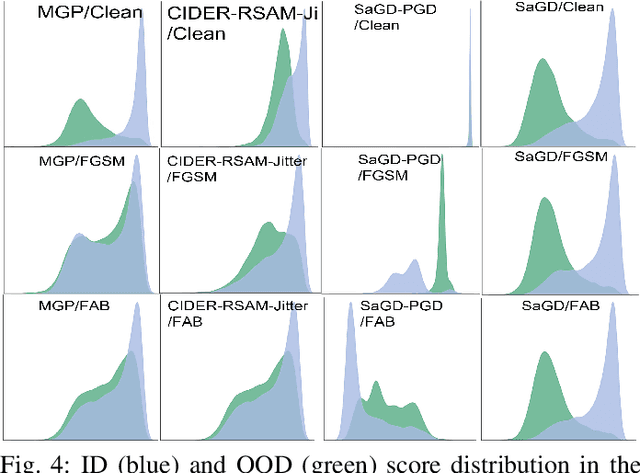
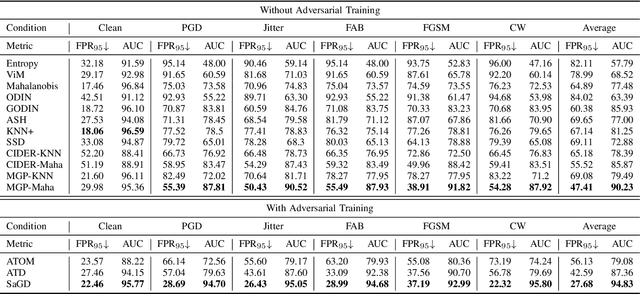
Abstract:Out-of-distribution (OOD) detection ensures safe and reliable model deployment. Contemporary OOD algorithms using geometry projection can detect OOD or adversarial samples from clean in-distribution (ID) samples. However, this setting regards adversarial ID samples as OOD, leading to incorrect OOD predictions. Existing efforts on OOD detection with ID and OOD data under attacks are minimal. In this paper, we develop a robust OOD detection method that distinguishes adversarial ID samples from OOD ones. The sharp loss landscape created by adversarial training hinders model convergence, impacting the latent embedding quality for OOD score calculation. Therefore, we introduce a {\bf Sharpness-aware Geometric Defense (SaGD)} framework to smooth out the rugged adversarial loss landscape in the projected latent geometry. Enhanced geometric embedding convergence enables accurate ID data characterization, benefiting OOD detection against adversarial attacks. We use Jitter-based perturbation in adversarial training to extend the defense ability against unseen attacks. Our SaGD framework significantly improves FPR and AUC over the state-of-the-art defense approaches in differentiating CIFAR-100 from six other OOD datasets under various attacks. We further examine the effects of perturbations at various adversarial training levels, revealing the relationship between the sharp loss landscape and adversarial OOD detection.
SeamlessEdit: Background Noise Aware Zero-Shot Speech Editing with in-Context Enhancement
May 20, 2025Abstract:With the fast development of zero-shot text-to-speech technologies, it is possible to generate high-quality speech signals that are indistinguishable from the real ones. Speech editing, including speech insertion and replacement, appeals to researchers due to its potential applications. However, existing studies only considered clean speech scenarios. In real-world applications, the existence of environmental noise could significantly degrade the quality of the generation. In this study, we propose a noise-resilient speech editing framework, SeamlessEdit, for noisy speech editing. SeamlessEdit adopts a frequency-band-aware noise suppression module and an in-content refinement strategy. It can well address the scenario where the frequency bands of voice and background noise are not separated. The proposed SeamlessEdit framework outperforms state-of-the-art approaches in multiple quantitative and qualitative evaluations.
Who Brings the Frisbee: Probing Hidden Hallucination Factors in Large Vision-Language Model via Causality Analysis
Dec 04, 2024
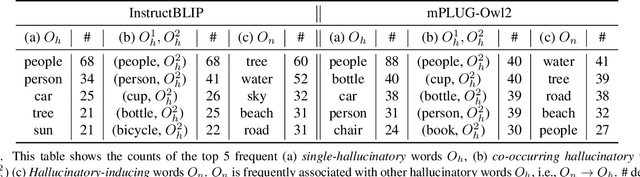
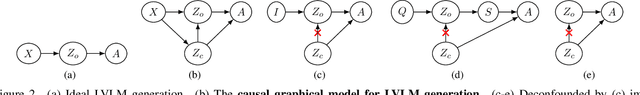
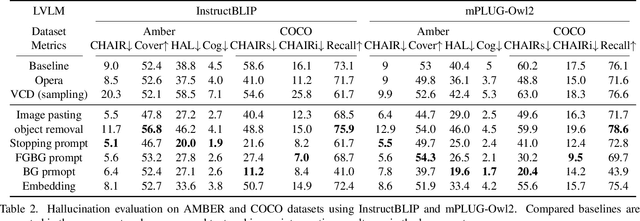
Abstract:Recent advancements in large vision-language models (LVLM) have significantly enhanced their ability to comprehend visual inputs alongside natural language. However, a major challenge in their real-world application is hallucination, where LVLMs generate non-existent visual elements, eroding user trust. The underlying mechanism driving this multimodal hallucination is poorly understood. Minimal research has illuminated whether contexts such as sky, tree, or grass field involve the LVLM in hallucinating a frisbee. We hypothesize that hidden factors, such as objects, contexts, and semantic foreground-background structures, induce hallucination. This study proposes a novel causal approach: a hallucination probing system to identify these hidden factors. By analyzing the causality between images, text prompts, and network saliency, we systematically explore interventions to block these factors. Our experimental findings show that a straightforward technique based on our analysis can significantly reduce hallucinations. Additionally, our analyses indicate the potential to edit network internals to minimize hallucinated outputs.
Improving Robustness and Clinical Applicability of Respiratory Sound Classification via Audio Enhancement
Jul 18, 2024Abstract:Deep learning techniques have shown promising results in the automatic classification of respiratory sounds. However, accurately distinguishing these sounds in real-world noisy conditions poses challenges for clinical deployment. Additionally, predicting signals with only background noise could undermine user trust in the system. In this study, we propose an audio enhancement (AE) pipeline as a pre-processing step before respiratory sound classification, aiming to improve performance in noisy environments. Multiple experiments were conducted using different audio enhancement model structures, demonstrating improved classification performance compared to the baseline method of noise injection data augmentation. Specifically, the integration of the AE pipeline resulted in a 2.59% increase in the ICBHI classification score on the ICBHI respiratory sound dataset and a 2.51% improvement on our recently collected Formosa Archive of Breath Sounds (FABS) in multi-class noisy scenarios. Furthermore, a physician validation study assessed the clinical utility of our system. Quantitative analysis revealed enhancements in efficiency, diagnostic confidence, and trust during model-assisted diagnosis with our system compared to raw noisy recordings. Workflows integrating enhanced audio led to an 11.61% increase in diagnostic sensitivity and facilitated high-confidence diagnoses. Our findings demonstrate that incorporating an audio enhancement algorithm significantly enhances robustness and clinical utility.
A Comprehensive Review of Machine Learning Advances on Data Change: A Cross-Field Perspective
Feb 20, 2024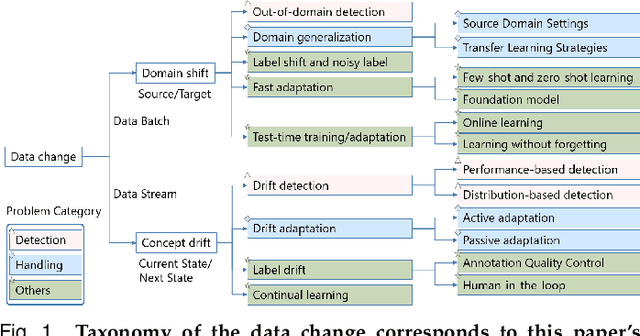
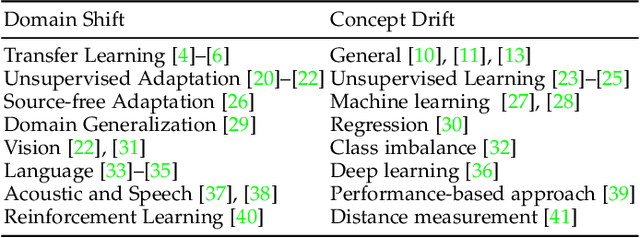
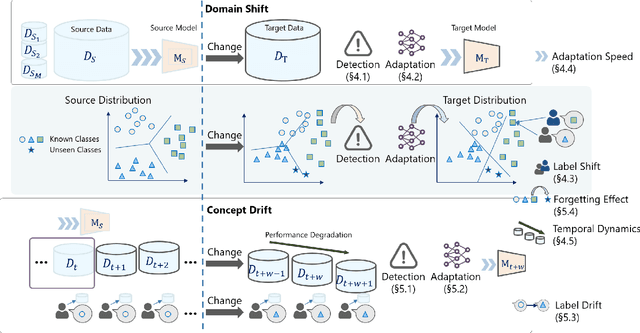
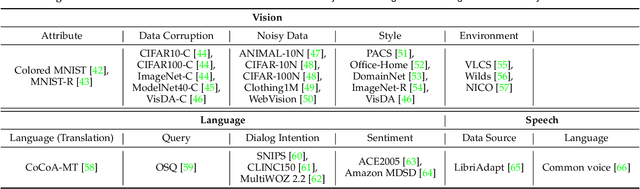
Abstract:Recent artificial intelligence (AI) technologies show remarkable evolution in various academic fields and industries. However, in the real world, dynamic data lead to principal challenges for deploying AI models. An unexpected data change brings about severe performance degradation in AI models. We identify two major related research fields, domain shift and concept drift according to the setting of the data change. Although these two popular research fields aim to solve distribution shift and non-stationary data stream problems, the underlying properties remain similar which also encourages similar technical approaches. In this review, we regroup domain shift and concept drift into a single research problem, namely the data change problem, with a systematic overview of state-of-the-art methods in the two research fields. We propose a three-phase problem categorization scheme to link the key ideas in the two technical fields. We thus provide a novel scope for researchers to explore contemporary technical strategies, learn industrial applications, and identify future directions for addressing data change challenges.
Improving Limited Supervised Foot Ulcer Segmentation Using Cross-Domain Augmentation
Jan 16, 2024Abstract:Diabetic foot ulcers pose health risks, including higher morbidity, mortality, and amputation rates. Monitoring wound areas is crucial for proper care, but manual segmentation is subjective due to complex wound features and background variation. Expert annotations are costly and time-intensive, thus hampering large dataset creation. Existing segmentation models relying on extensive annotations are impractical in real-world scenarios with limited annotated data. In this paper, we propose a cross-domain augmentation method named TransMix that combines Augmented Global Pre-training AGP and Localized CutMix Fine-tuning LCF to enrich wound segmentation data for model learning. TransMix can effectively improve the foot ulcer segmentation model training by leveraging other dermatology datasets not on ulcer skins or wounds. AGP effectively increases the overall image variability, while LCF increases the diversity of wound regions. Experimental results show that TransMix increases the variability of wound regions and substantially improves the Dice score for models trained with only 40 annotated images under various proportions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge