Jeffrey C. Grossman
A cloud platform for automating and sharing analysis of raw simulation data from high throughput polymer molecular dynamics simulations
Aug 02, 2022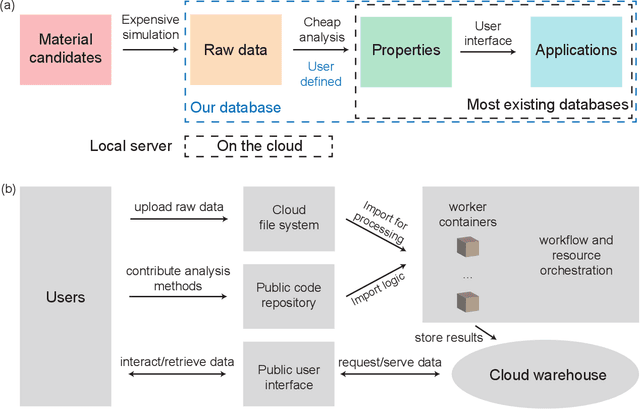
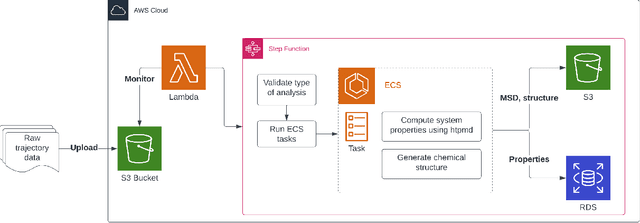
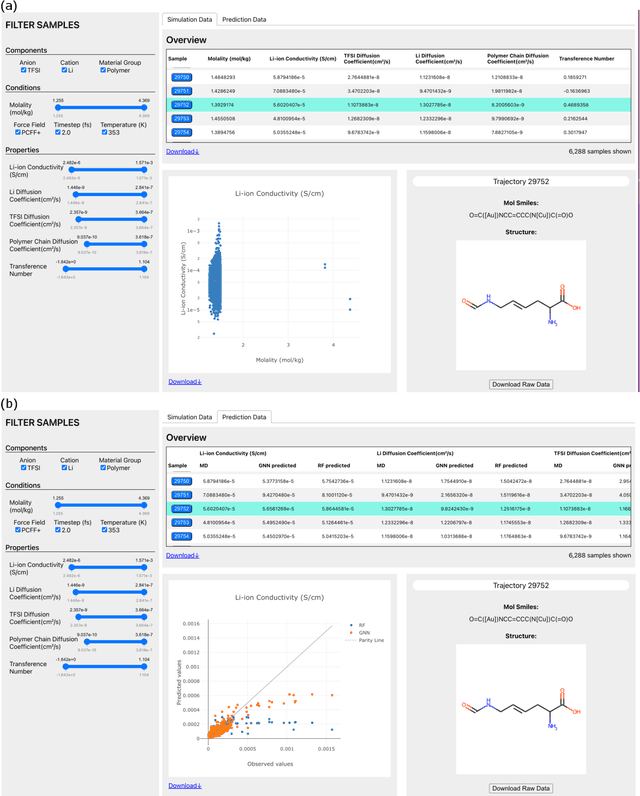
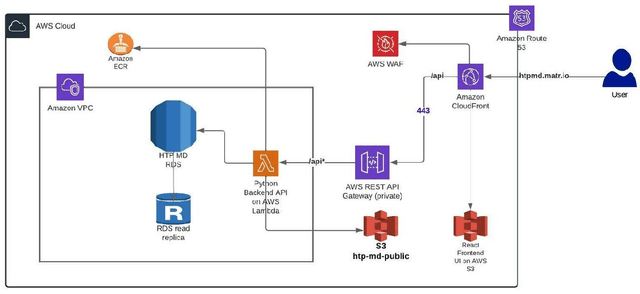
Abstract:Open material databases storing hundreds of thousands of material structures and their corresponding properties have become the cornerstone of modern computational materials science. Yet, the raw outputs of the simulations, such as the trajectories from molecular dynamics simulations and charge densities from density functional theory calculations, are generally not shared due to their huge size. In this work, we describe a cloud-based platform to facilitate the sharing of raw data and enable the fast post-processing in the cloud to extract new properties defined by the user. As an initial demonstration, our database currently includes 6286 molecular dynamics trajectories for amorphous polymer electrolytes and 5.7 terabytes of data. We create a public analysis library at https://github.com/TRI-AMDD/htp_md to extract multiple properties from the raw data, using both expert designed functions and machine learning models. The analysis is run automatically with computation in the cloud, and results then populate a database that can be accessed publicly. Our platform encourages users to contribute both new trajectory data and analysis functions via public interfaces. Newly analyzed properties will be incorporated into the database. Finally, we create a front-end user interface at https://www.htpmd.matr.io for browsing and visualization of our data. We envision the platform to be a new way of sharing raw data and new insights for the computational materials science community.
Accelerating the screening of amorphous polymer electrolytes by learning to reduce random and systematic errors in molecular dynamics simulations
Jan 13, 2021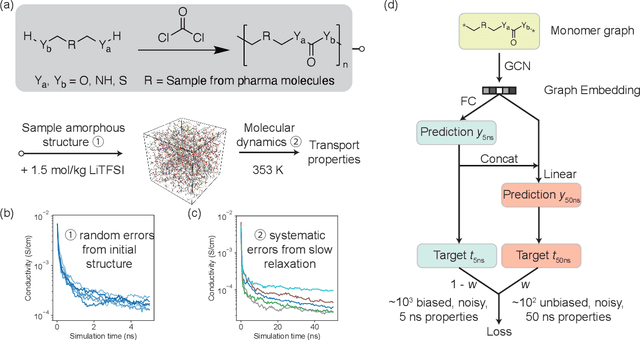
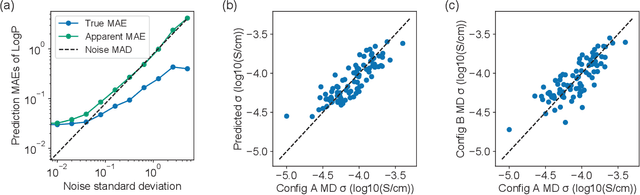
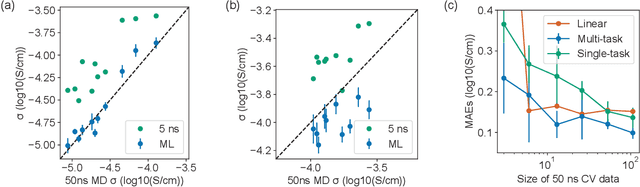
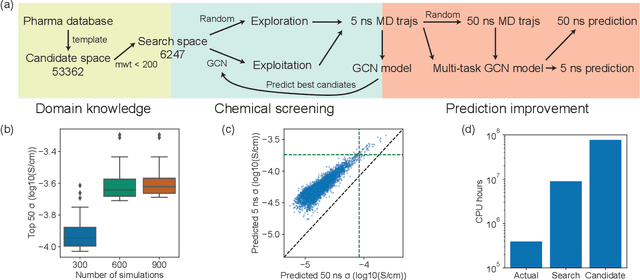
Abstract:Machine learning has been widely adopted to accelerate the screening of materials. Most existing studies implicitly assume that the training data are generated through a deterministic, unbiased process, but this assumption might not hold for the simulation of some complex materials. In this work, we aim to screen amorphous polymer electrolytes which are promising candidates for the next generation lithium-ion battery technology but extremely expensive to simulate due to their structural complexity. We demonstrate that a multi-task graph neural network can learn from a large amount of noisy, biased data and a small number of unbiased data and reduce both random and systematic errors in predicting the transport properties of polymer electrolytes. This observation allows us to achieve accurate predictions on the properties of complex materials by learning to reduce errors in the training data, instead of running repetitive, expensive simulations which is conventionally used to reduce simulation errors. With this approach, we screen a space of 6247 polymer electrolytes, orders of magnitude larger than previous computational studies. We also find a good extrapolation performance to the top polymers from a larger space of 53362 polymers and 31 experimentally-realized polymers. The strategy employed in this work may be applicable to a broad class of material discovery problems that involve the simulation of complex, amorphous materials.
Graph Dynamical Networks: Unsupervised Learning of Atomic Scale Dynamics in Materials
Feb 18, 2019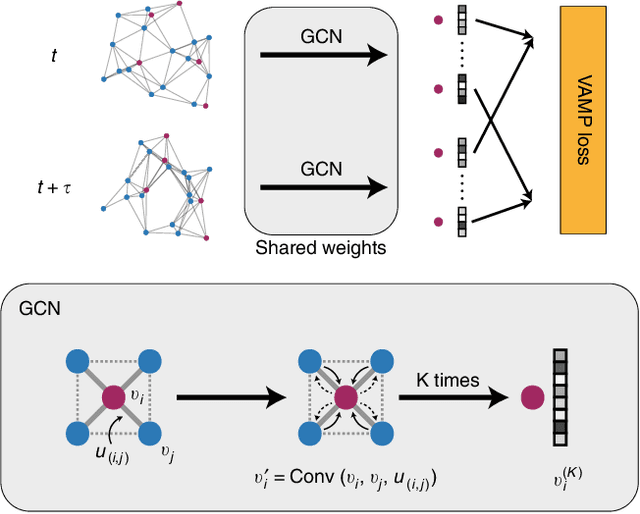
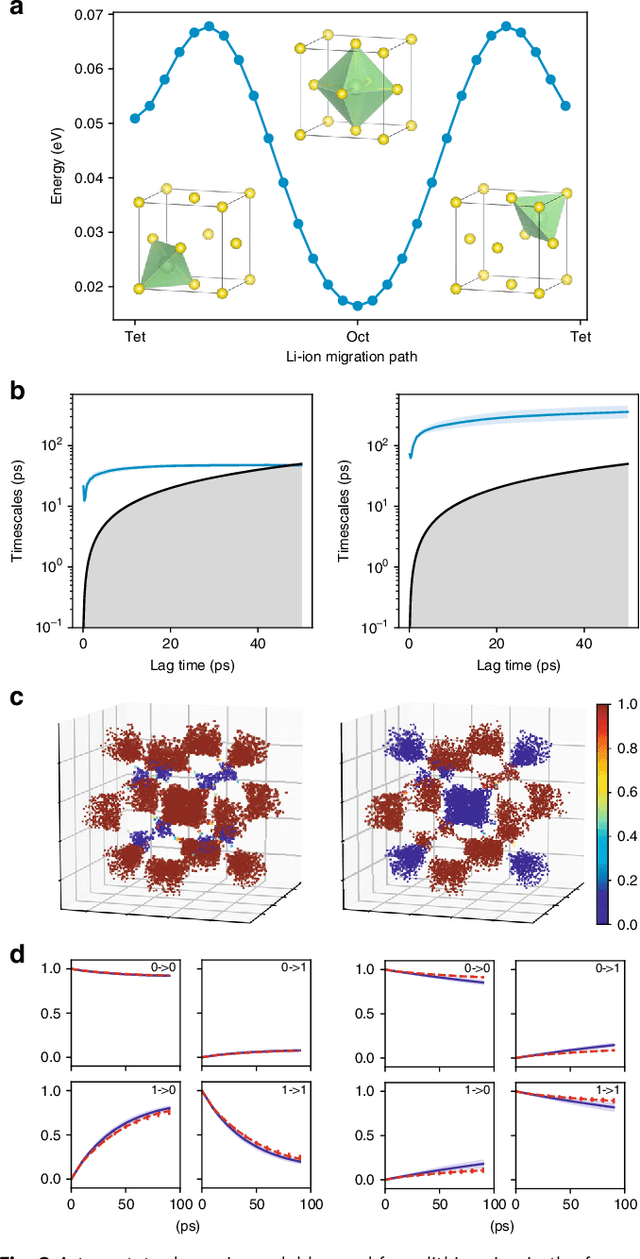
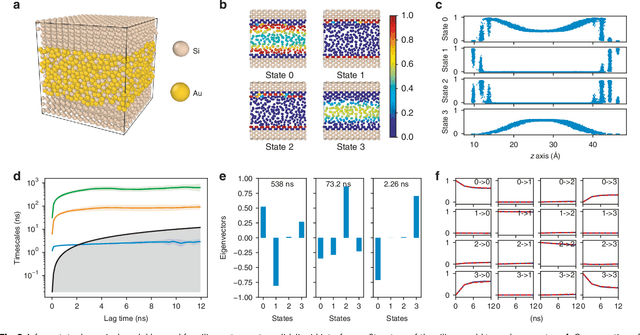
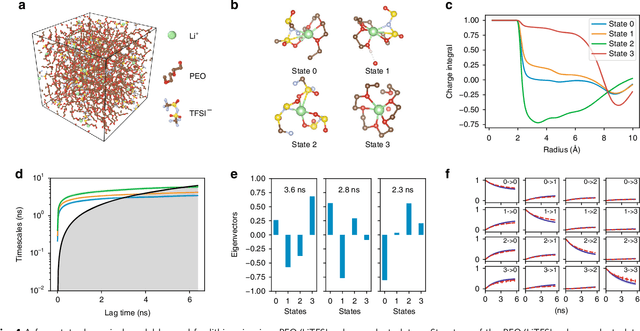
Abstract:Understanding the dynamical processes that govern the performance of functional materials is essential for the design of next generation materials to tackle global energy and environmental challenges. Many of these processes involve the dynamics of individual atoms or small molecules in condensed phases, e.g. lithium ions in electrolytes, water molecules in membranes, molten atoms at interfaces, etc., which are difficult to understand due to the complexity of local environments. In this work, we develop graph dynamical networks, an unsupervised learning approach for understanding atomic scale dynamics in arbitrary phases and environments from molecular dynamics simulations. We demonstrate that learning the local dynamics of atoms can be significantly easier than the global dynamics of the entire material system using a toy system. We also apply the method to learn the dynamics of two different systems -- silicon atoms at liquid-solid interfaces, and lithium ions in amorphous polymer electrolytes, and show that our approach gains important dynamical information that is otherwise difficult to obtain. With the large amounts of molecular dynamics data generated everyday in nearly every aspect of materials design, this approach provides a broadly useful, automated tool to understand atomic scale dynamics in material systems.
Hierarchical Visualization of Materials Space with Graph Convolutional Neural Networks
Sep 18, 2018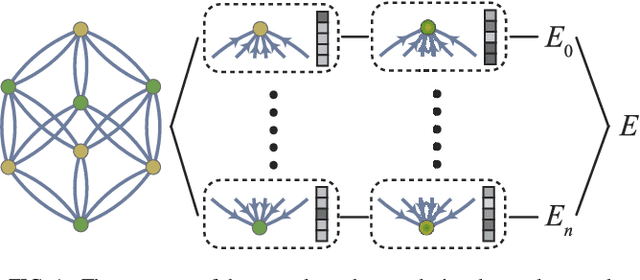
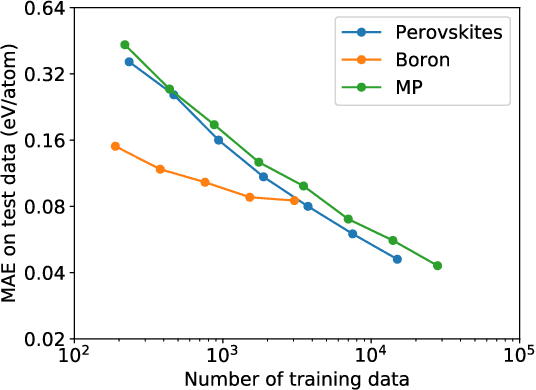
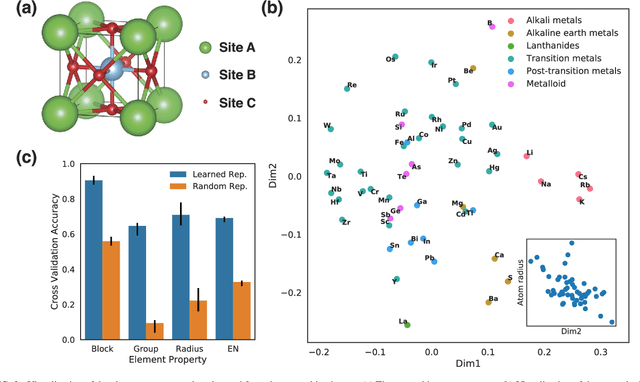
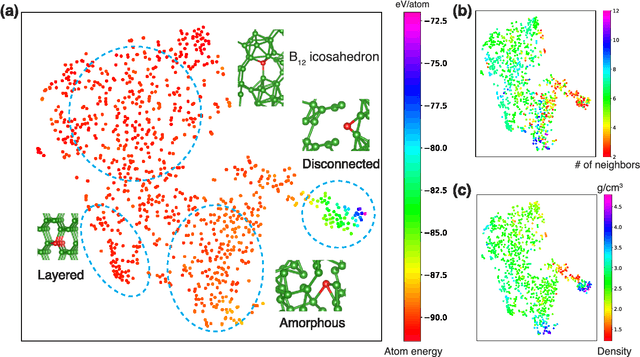
Abstract:The combination of high throughput computation and machine learning has led to a new paradigm in materials design by allowing for the direct screening of vast portions of structural, chemical, and property space. The use of these powerful techniques leads to the generation of enormous amounts of data, which in turn calls for new techniques to efficiently explore and visualize the materials space to help identify underlying patterns. In this work, we develop a unified framework to hierarchically visualize the compositional and structural similarities between materials in an arbitrary material space with representations learned from different layers of graph convolutional neural networks. We demonstrate the potential for such a visualization approach by showing that patterns emerge automatically that reflect similarities at different scales in three representative classes of materials: perovskites, elemental boron, and general inorganic crystals, covering material spaces of different compositions, structures, and both. For perovskites, elemental similarities are learned that reflects multiple aspects of atom properties. For elemental boron, structural motifs emerge automatically showing characteristic boron local environments. For inorganic crystals, the similarity and stability of local coordination environments are shown combining different center and neighbor atoms. The method could help transition to a data-centered exploration of materials space in automated materials design.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge