Graph Dynamical Networks: Unsupervised Learning of Atomic Scale Dynamics in Materials
Paper and Code
Feb 18, 2019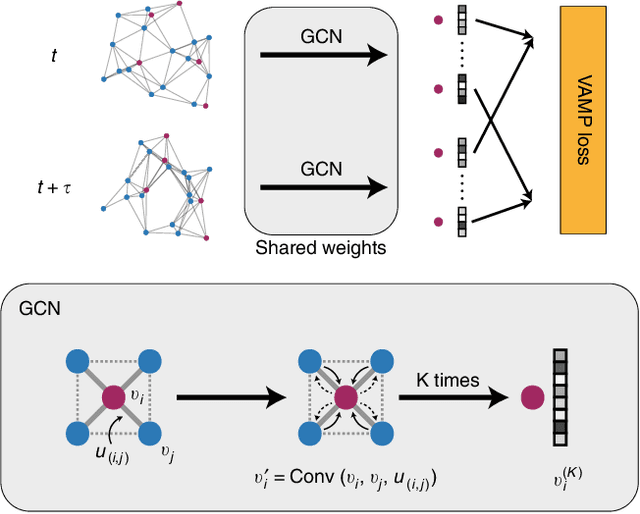
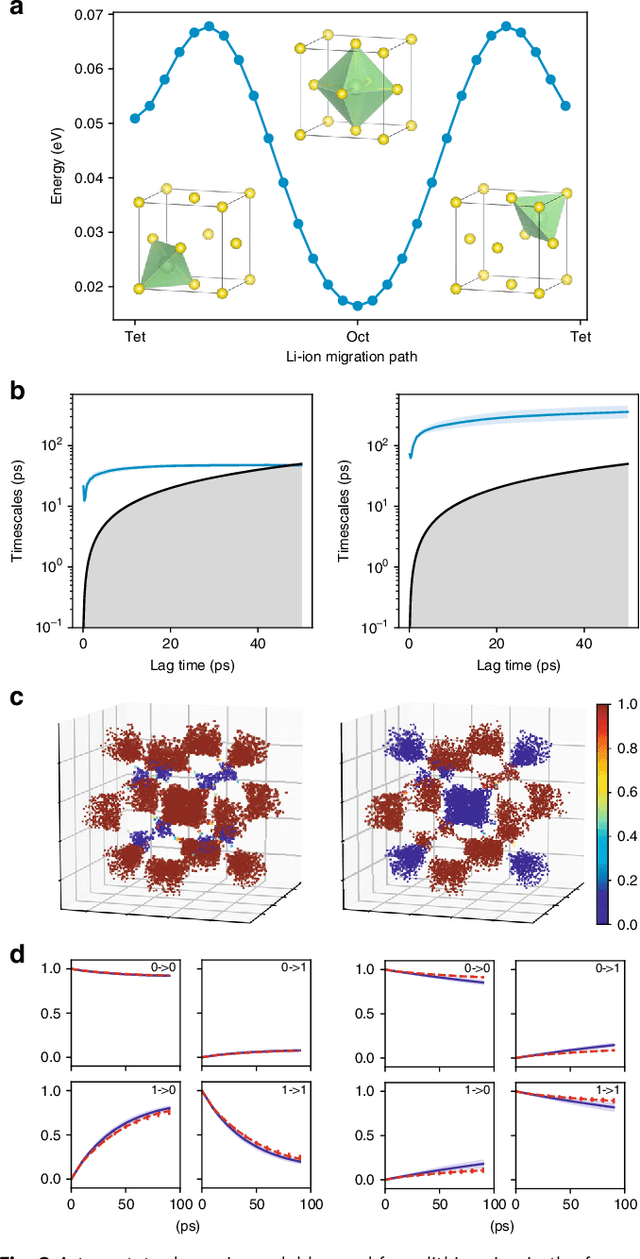
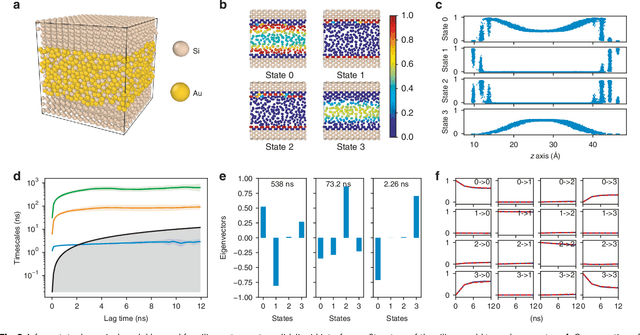
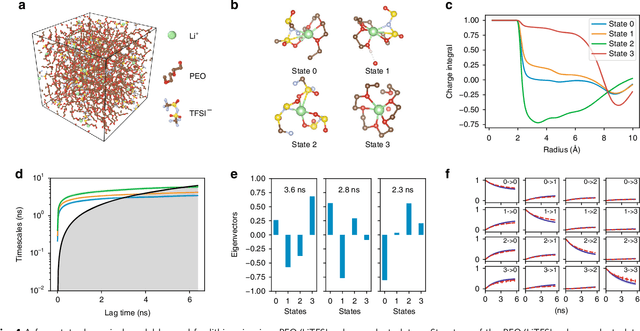
Understanding the dynamical processes that govern the performance of functional materials is essential for the design of next generation materials to tackle global energy and environmental challenges. Many of these processes involve the dynamics of individual atoms or small molecules in condensed phases, e.g. lithium ions in electrolytes, water molecules in membranes, molten atoms at interfaces, etc., which are difficult to understand due to the complexity of local environments. In this work, we develop graph dynamical networks, an unsupervised learning approach for understanding atomic scale dynamics in arbitrary phases and environments from molecular dynamics simulations. We demonstrate that learning the local dynamics of atoms can be significantly easier than the global dynamics of the entire material system using a toy system. We also apply the method to learn the dynamics of two different systems -- silicon atoms at liquid-solid interfaces, and lithium ions in amorphous polymer electrolytes, and show that our approach gains important dynamical information that is otherwise difficult to obtain. With the large amounts of molecular dynamics data generated everyday in nearly every aspect of materials design, this approach provides a broadly useful, automated tool to understand atomic scale dynamics in material systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge