Jayanthi Sivaswamy
A metric to compare the anatomy variation between image time series
Feb 23, 2023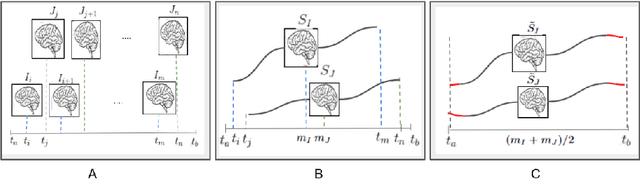
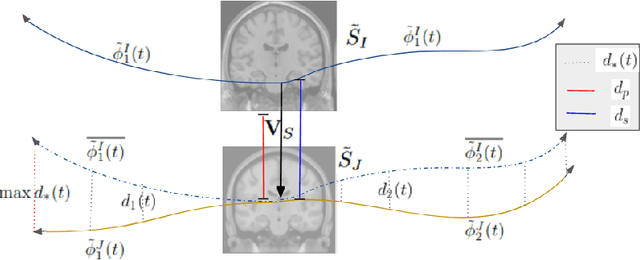
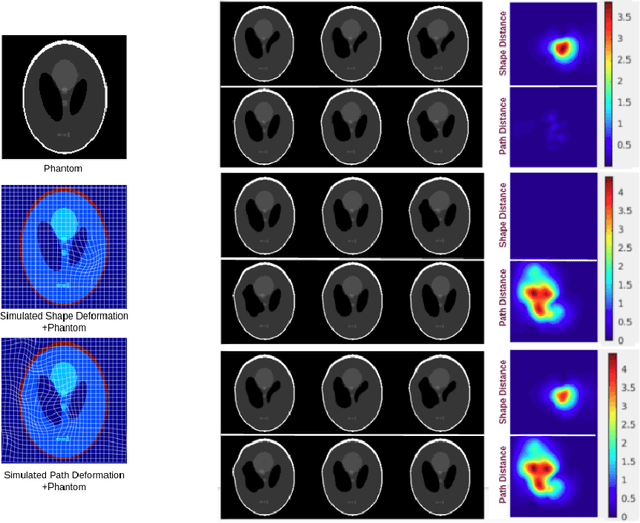
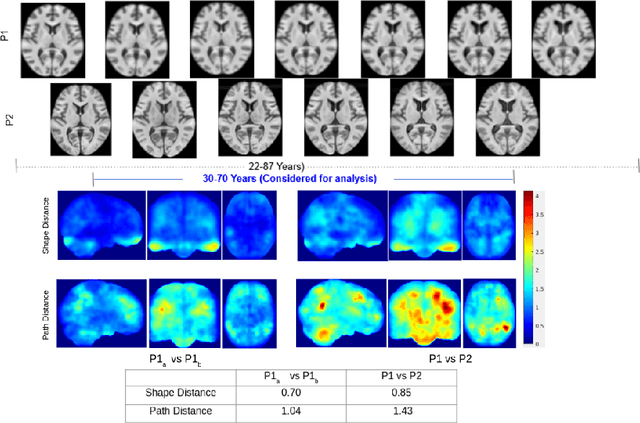
Abstract:Biological processes like growth, aging, and disease progression are generally studied with follow-up scans taken at different time points, i.e., with image time series (TS) based analysis. Comparison between TS representing a biological process of two individuals/populations is of interest. A metric to quantify the difference between TS is desirable for such a comparison. The two TS represent the evolution of two different subject/population average anatomies through two paths. A method to untangle and quantify the path and inter-subject anatomy(shape) difference between the TS is presented in this paper. The proposed metric is a generalized version of Fr\'echet distance designed to compare curves. The proposed method is evaluated with simulated and adult and fetal neuro templates. Results show that the metric is able to separate and quantify the path and shape differences between TS.
Sub-cortical structure segmentation database for young population
Nov 10, 2021



Abstract:Segmentation of sub-cortical structures from MRI scans is of interest in many neurological diagnosis. Since this is a laborious task machine learning and specifically deep learning (DL) methods have become explored. The structural complexity of the brain demands a large, high quality segmentation dataset to develop good DL-based solutions for sub-cortical structure segmentation. Towards this, we are releasing a set of 114, 1.5 Tesla, T1 MRI scans with manual delineations for 14 sub-cortical structures. The scans in the dataset were acquired from healthy young (21-30 years) subjects ( 58 male and 56 female) and all the structures are manually delineated by experienced radiology experts. Segmentation experiments have been conducted with this dataset and results demonstrate that accurate results can be obtained with deep-learning methods. Our sub-cortical structure segmentation dataset, Indian Brain Segmentation Dataset (IBSD) is made openly available at \url{https://doi.org/10.5281/zenodo.5656776}.
A Diffeomorphic Aging Model for Adult Human Brain from Cross-Sectional Data
Jun 28, 2021



Abstract:Normative aging trends of the brain can serve as an important reference in the assessment of neurological structural disorders. Such models are typically developed from longitudinal brain image data -- follow-up data of the same subject over different time points. In practice, obtaining such longitudinal data is difficult. We propose a method to develop an aging model for a given population, in the absence of longitudinal data, by using images from different subjects at different time points, the so-called cross-sectional data. We define an aging model as a diffeomorphic deformation on a structural template derived from the data and propose a method that develops topology preserving aging model close to natural aging. The proposed model is successfully validated on two public cross-sectional datasets which provide templates constructed from different sets of subjects at different age points.
A Clinically Inspired Approach for Melanoma classification
Jun 15, 2021


Abstract:Melanoma is a leading cause of deaths due to skin cancer deaths and hence, early and effective diagnosis of melanoma is of interest. Current approaches for automated diagnosis of melanoma either use pattern recognition or analytical recognition like ABCDE (asymmetry, border, color, diameter and evolving) criterion. In practice however, a differential approach wherein outliers (ugly duckling) are detected and used to evaluate nevi/lesions. Incorporation of differential recognition in Computer Aided Diagnosis (CAD) systems has not been explored but can be beneficial as it can provide a clinical justification for the derived decision. We present a method for identifying and quantifying ugly ducklings by performing Intra-Patient Comparative Analysis (IPCA) of neighboring nevi. This is then incorporated in a CAD system design for melanoma detection. This design ensures flexibility to handle cases where IPCA is not possible. Our experiments on a public dataset show that the outlier information helps boost the sensitivity of detection by at least 4.1 % and specificity by 4.0 % to 8.9 %, depending on the use of a strong (EfficientNet) or moderately strong (VGG or ResNet) classifier.
Self-Supervised Learning for Segmentation
Jan 14, 2021



Abstract:Self-supervised learning is emerging as an effective substitute for transfer learning from large datasets. In this work, we use kidney segmentation to explore this idea. The anatomical asymmetry of kidneys is leveraged to define an effective proxy task for kidney segmentation via self-supervised learning. A siamese convolutional neural network (CNN) is used to classify a given pair of kidney sections from CT volumes as being kidneys of the same or different sides. This knowledge is then transferred for the segmentation of kidneys using another deep CNN using one branch of the siamese CNN as the encoder for the segmentation network. Evaluation results on a publicly available dataset containing computed tomography (CT) scans of the abdominal region shows that a boost in performance and fast convergence can be had relative to a network trained conventionally from scratch. This is notable given that no additional data/expensive annotations or augmentation were used in training.
A method for large diffeomorphic registration via broken geodesics
Jan 03, 2021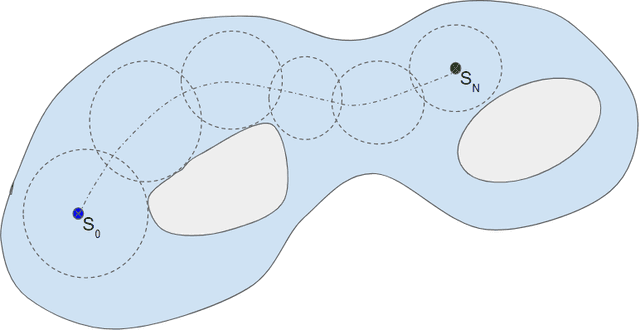



Abstract:Anatomical variabilities seen in longitudinal data or inter-subject data is usually described by the underlying deformation, captured by non-rigid registration of these images. Stationary Velocity Field (SVF) based non-rigid registration algorithms are widely used for registration. SVF based methods form a metric-free framework which captures a finite dimensional submanifold of deformations embedded in the infinite dimensional smooth manifold of diffeomorphisms. However, these methods cover only a limited degree of deformations. In this paper, we address this limitation and define an approximate metric space for the manifold of diffeomorphisms $\mathcal{G}$. We propose a method to break down the large deformation into finite compositions of small deformations. This results in a broken geodesic path on $\mathcal{G}$ and its length now forms an approximate registration metric. We illustrate the method using a simple, intensity-based, log-demon implementation. Validation results of the proposed method show that it can capture large and complex deformations while producing qualitatively better results than the state-of-the-art methods. The results also demonstrate that the proposed registration metric is a good indicator of the degree of deformation.
Explainable Disease Classification via weakly-supervised segmentation
Aug 24, 2020

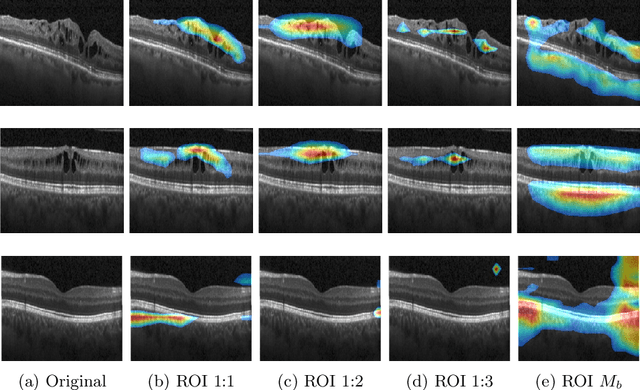

Abstract:Deep learning based approaches to Computer Aided Diagnosis (CAD) typically pose the problem as an image classification (Normal or Abnormal) problem. These systems achieve high to very high accuracy in specific disease detection for which they are trained but lack in terms of an explanation for the provided decision/classification result. The activation maps which correspond to decisions do not correlate well with regions of interest for specific diseases. This paper examines this problem and proposes an approach which mimics the clinical practice of looking for an evidence prior to diagnosis. A CAD model is learnt using a mixed set of information: class labels for the entire training set of images plus a rough localisation of suspect regions as an extra input for a smaller subset of training images for guiding the learning. The proposed approach is illustrated with detection of diabetic macular edema (DME) from OCT slices. Results of testing on on a large public dataset show that with just a third of images with roughly segmented fluid filled regions, the classification accuracy is on par with state of the art methods while providing a good explanation in the form of anatomically accurate heatmap /region of interest. The proposed solution is then adapted to Breast Cancer detection from mammographic images. Good evaluation results on public datasets underscores the generalisability of the proposed solution.
Image Segmentation Using Hybrid Representations
Apr 15, 2020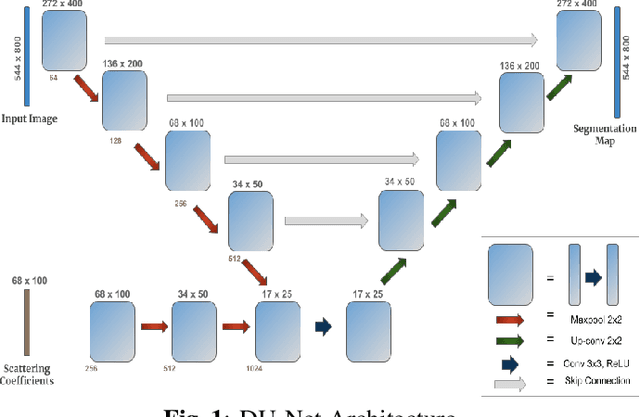
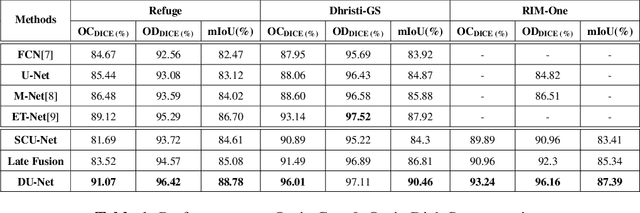
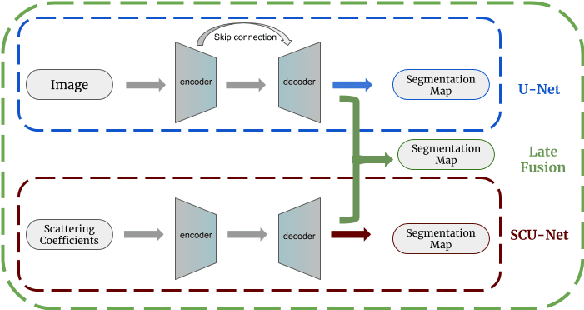
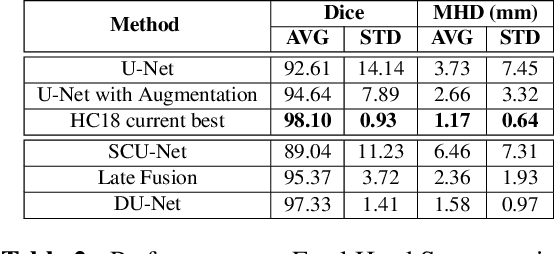
Abstract:This work explores a hybrid approach to segmentation as an alternative to a purely data-driven approach. We introduce an end-to-end U-Net based network called DU-Net, which uses additional frequency preserving features, namely the Scattering Coefficients (SC), for medical image segmentation. SC are translation invariant and Lipschitz continuous to deformations which help DU-Net outperform other conventional CNN counterparts on four datasets and two segmentation tasks: Optic Disc and Optic Cup in color fundus images and fetal Head in ultrasound images. The proposed method shows remarkable improvement over the basic U-Net with performance competitive to state-of-the-art methods. The results indicate that it is possible to use a lighter network trained with fewer images (without any augmentation) to attain good segmentation results.
FPD-M-net: Fingerprint Image Denoising and Inpainting Using M-Net Based Convolutional Neural Networks
Dec 26, 2018



Abstract:The fingerprint is a common biometric used for authentication and verification of an individual. These images are degraded when fingers are wet, dirty, dry or wounded and due to the failure of the sensors, etc. The extraction of the fingerprint from a degraded image requires denoising and inpainting. We propose to address these problems with an end-to-end trainable Convolutional Neural Network based architecture called FPD-M-net, by posing the fingerprint denoising and inpainting problem as a segmentation (foreground) task. Our architecture is based on the M-net with a change: structure similarity loss function, used for better extraction of the fingerprint from the noisy background. Our method outperforms the baseline method and achieves an overall 3rd rank in the Chalearn LAP Inpainting Competition Track 3 - Fingerprint Denoising and Inpainting, ECCV 2018
To Learn or Not to Learn Features for Deformable Registration?
Jul 06, 2018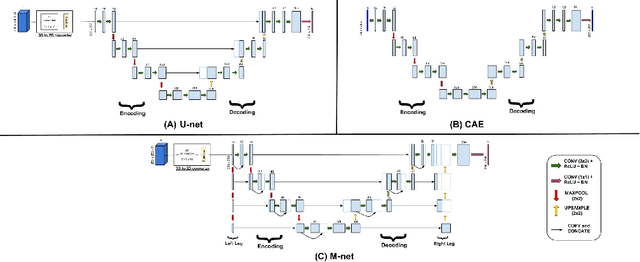

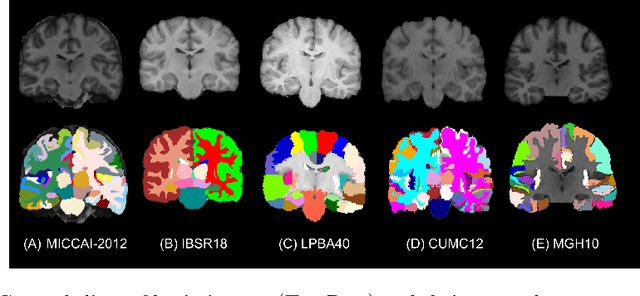
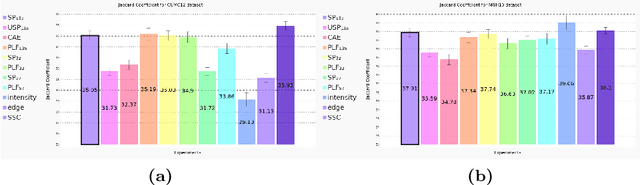
Abstract:Feature-based registration has been popular with a variety of features ranging from voxel intensity to Self-Similarity Context (SSC). In this paper, we examine the question on how features learnt using various Deep Learning (DL) frameworks can be used for deformable registration and whether this feature learning is necessary or not. We investigate the use of features learned by different DL methods in the current state-of-the-art discrete registration framework and analyze its performance on 2 publicly available datasets. We draw insights into the type of DL framework useful for feature learning and the impact, if any, of the complexity of different DL models and brain parcellation methods on the performance of discrete registration. Our results indicate that the registration performance with DL features and SSC are comparable and stable across datasets whereas this does not hold for low level features.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge