Ruchi Chauhan
Exploring Genetic-histologic Relationships in Breast Cancer
Mar 15, 2021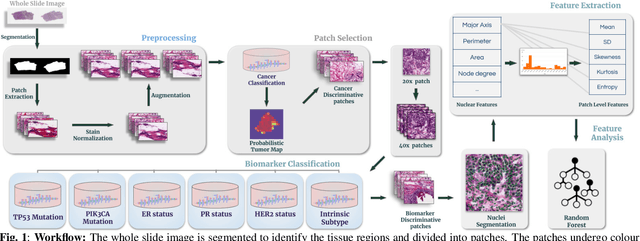
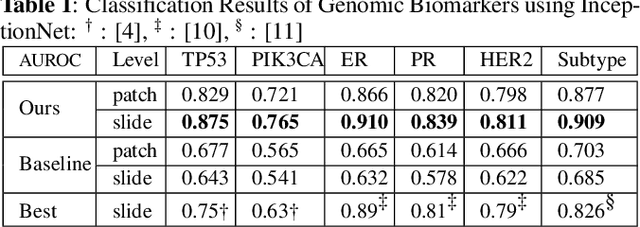
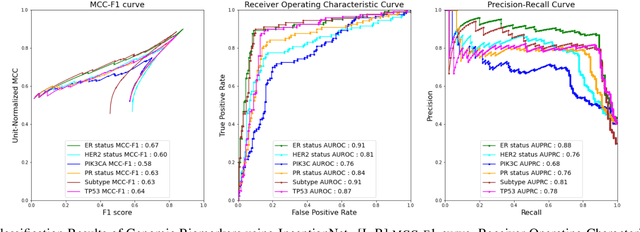
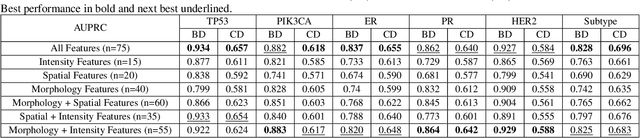
Abstract:The advent of digital pathology presents opportunities for computer vision for fast, accurate, and objective solutions for histopathological images and aid in knowledge discovery. This work uses deep learning to predict genomic biomarkers - TP53 mutation, PIK3CA mutation, ER status, PR status, HER2 status, and intrinsic subtypes, from breast cancer histopathology images. Furthermore, we attempt to understand the underlying morphology as to how these genomic biomarkers manifest in images. Since gene sequencing is expensive, not always available, or even feasible, predicting these biomarkers from images would help in diagnosis, prognosis, and effective treatment planning. We outperform the existing works with a minimum improvement of 0.02 and a maximum of 0.13 AUROC scores across all tasks. We also gain insights that can serve as hypotheses for further experimentations, including the presence of lymphocytes and karyorrhexis. Moreover, our fully automated workflow can be extended to other tasks across other cancer subtypes.
Image Segmentation Using Hybrid Representations
Apr 15, 2020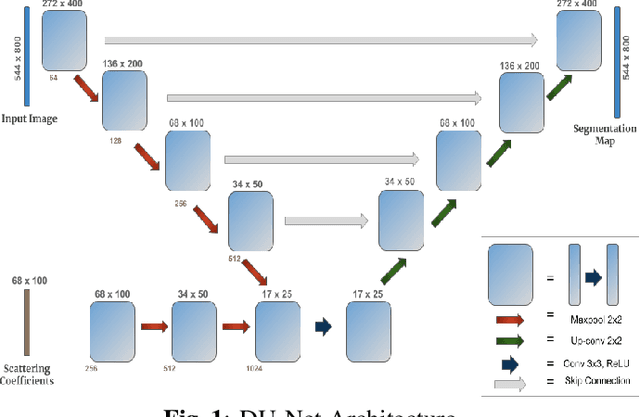
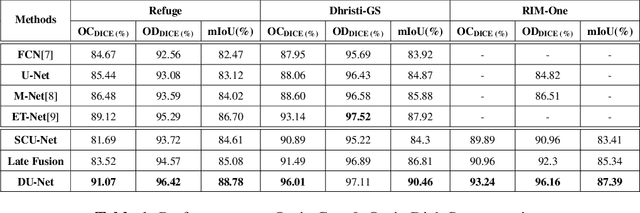
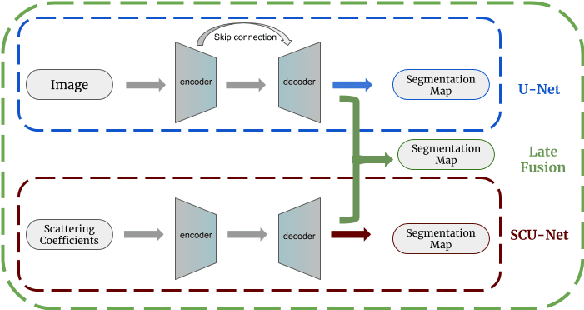
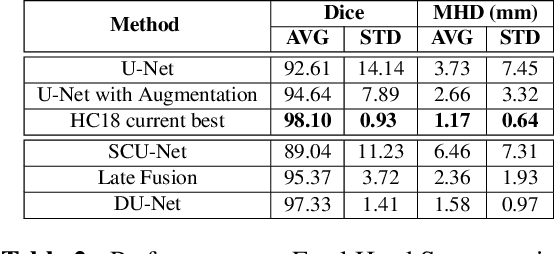
Abstract:This work explores a hybrid approach to segmentation as an alternative to a purely data-driven approach. We introduce an end-to-end U-Net based network called DU-Net, which uses additional frequency preserving features, namely the Scattering Coefficients (SC), for medical image segmentation. SC are translation invariant and Lipschitz continuous to deformations which help DU-Net outperform other conventional CNN counterparts on four datasets and two segmentation tasks: Optic Disc and Optic Cup in color fundus images and fetal Head in ultrasound images. The proposed method shows remarkable improvement over the basic U-Net with performance competitive to state-of-the-art methods. The results indicate that it is possible to use a lighter network trained with fewer images (without any augmentation) to attain good segmentation results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge