Alphin J Thottupattu
A metric to compare the anatomy variation between image time series
Feb 23, 2023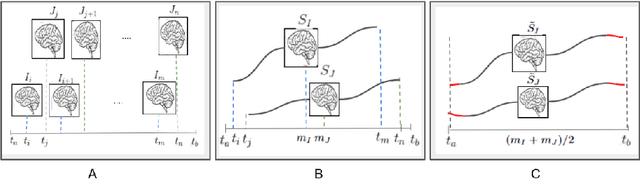
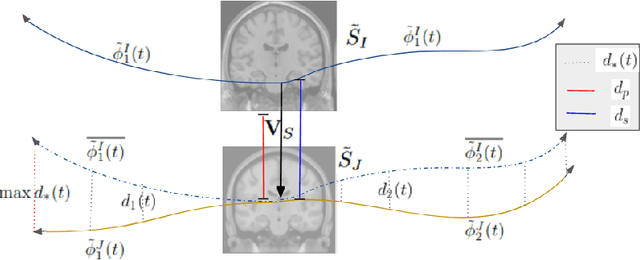
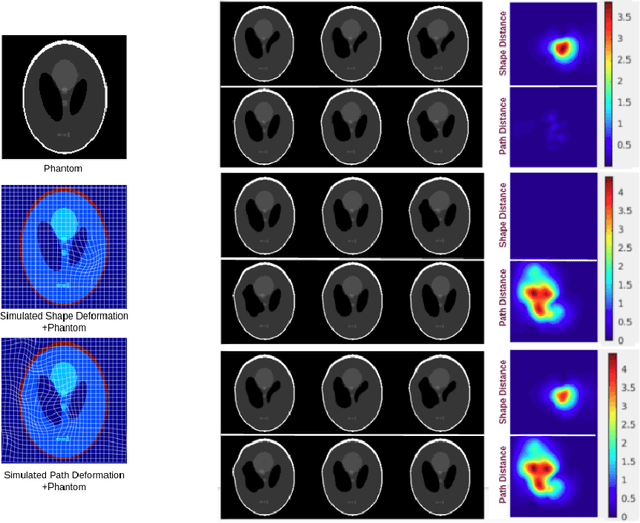
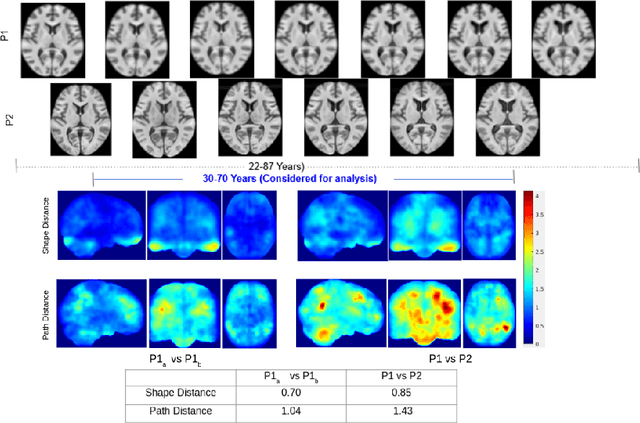
Abstract:Biological processes like growth, aging, and disease progression are generally studied with follow-up scans taken at different time points, i.e., with image time series (TS) based analysis. Comparison between TS representing a biological process of two individuals/populations is of interest. A metric to quantify the difference between TS is desirable for such a comparison. The two TS represent the evolution of two different subject/population average anatomies through two paths. A method to untangle and quantify the path and inter-subject anatomy(shape) difference between the TS is presented in this paper. The proposed metric is a generalized version of Fr\'echet distance designed to compare curves. The proposed method is evaluated with simulated and adult and fetal neuro templates. Results show that the metric is able to separate and quantify the path and shape differences between TS.
Sub-cortical structure segmentation database for young population
Nov 10, 2021



Abstract:Segmentation of sub-cortical structures from MRI scans is of interest in many neurological diagnosis. Since this is a laborious task machine learning and specifically deep learning (DL) methods have become explored. The structural complexity of the brain demands a large, high quality segmentation dataset to develop good DL-based solutions for sub-cortical structure segmentation. Towards this, we are releasing a set of 114, 1.5 Tesla, T1 MRI scans with manual delineations for 14 sub-cortical structures. The scans in the dataset were acquired from healthy young (21-30 years) subjects ( 58 male and 56 female) and all the structures are manually delineated by experienced radiology experts. Segmentation experiments have been conducted with this dataset and results demonstrate that accurate results can be obtained with deep-learning methods. Our sub-cortical structure segmentation dataset, Indian Brain Segmentation Dataset (IBSD) is made openly available at \url{https://doi.org/10.5281/zenodo.5656776}.
A Diffeomorphic Aging Model for Adult Human Brain from Cross-Sectional Data
Jun 28, 2021



Abstract:Normative aging trends of the brain can serve as an important reference in the assessment of neurological structural disorders. Such models are typically developed from longitudinal brain image data -- follow-up data of the same subject over different time points. In practice, obtaining such longitudinal data is difficult. We propose a method to develop an aging model for a given population, in the absence of longitudinal data, by using images from different subjects at different time points, the so-called cross-sectional data. We define an aging model as a diffeomorphic deformation on a structural template derived from the data and propose a method that develops topology preserving aging model close to natural aging. The proposed model is successfully validated on two public cross-sectional datasets which provide templates constructed from different sets of subjects at different age points.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge