Jason Portenoy
Bursting Scientific Filter Bubbles: Boosting Innovation via Novel Author Discovery
Sep 10, 2021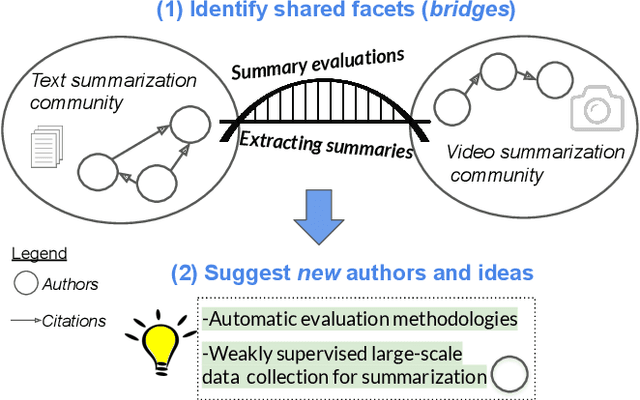
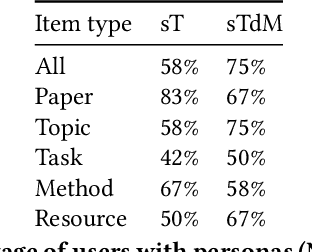
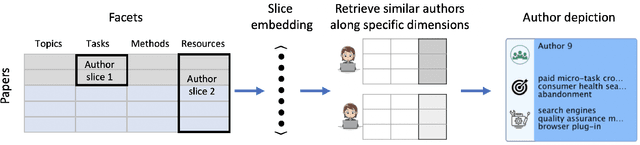
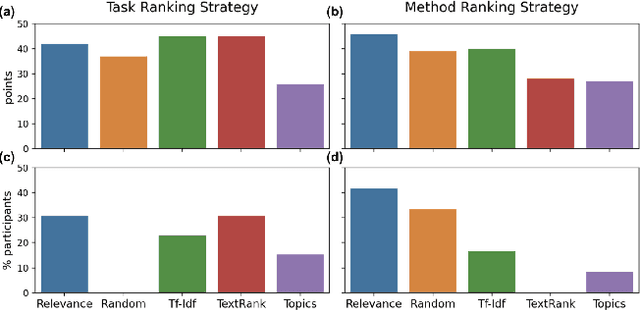
Abstract:Isolated silos of scientific research and the growing challenge of information overload limit awareness across the literature and hinder innovation. Algorithmic curation and recommendation, which often prioritize relevance, can further reinforce these informational "filter bubbles." In response, we describe Bridger, a system for facilitating discovery of scholars and their work, to explore design tradeoffs between relevant and novel recommendations. We construct a faceted representation of authors with information gleaned from their papers and inferred author personas, and use it to develop an approach that locates commonalities ("bridges") and contrasts between scientists -- retrieving partially similar authors rather than aiming for strict similarity. In studies with computer science researchers, this approach helps users discover authors considered useful for generating novel research directions, outperforming a state-of-art neural model. In addition to recommending new content, we also demonstrate an approach for displaying it in a manner that boosts researchers' ability to understand the work of authors with whom they are unfamiliar. Finally, our analysis reveals that Bridger connects authors who have different citation profiles, publish in different venues, and are more distant in social co-authorship networks, raising the prospect of bridging diverse communities and facilitating discovery.
SciSight: Combining faceted navigation and research group detection for COVID-19 exploratory scientific search
May 27, 2020
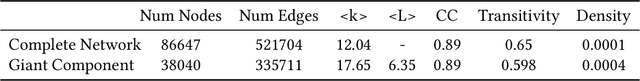


Abstract:The COVID-19 pandemic has sparked unprecedented mobilization of scientists, already generating thousands of new papers that join a litany of previous biomedical work in related areas. This deluge of information makes it hard for researchers to keep track of their own research area, let alone explore new directions. Standard search engines are designed primarily for targeted search and are not geared for discovery or making connections that are not obvious from reading individual papers. In this paper, we present our ongoing work on SciSight, a novel framework for exploratory search of COVID-19 research. Based on formative interviews with scientists and a review of existing tools, we build and integrate two key capabilities: first, exploring interactions between biomedical facets (e.g., proteins, genes, drugs, diseases, patient characteristics); and second, discovering groups of researchers and how they are connected. We extract entities using a language model pre-trained on several biomedical information extraction tasks, and enrich them with data from the Microsoft Academic Graph (MAG). To find research groups automatically, we use hierarchical clustering with overlap to allow authors, as they do, to belong to multiple groups. Finally, we introduce a novel presentation of these groups based on both topical and social affinities, allowing users to drill down from groups to papers to associations between entities, and update query suggestions on the fly with the goal of facilitating exploratory navigation. SciSight has thus far served over 10K users with over 30K page views and 13% returning users. Preliminary user interviews with biomedical researchers suggest that SciSight complements current approaches and helps find new and relevant knowledge.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge