Jan Aelterman
DYRECT Computed Tomography: DYnamic Reconstruction of Events on a Continuous Timescale
Nov 15, 2024



Abstract:Time-resolved high-resolution X-ray Computed Tomography (4D $\mu$CT) is an imaging technique that offers insight into the evolution of dynamic processes inside materials that are opaque to visible light. Conventional tomographic reconstruction techniques are based on recording a sequence of 3D images that represent the sample state at different moments in time. This frame-based approach limits the temporal resolution compared to dynamic radiography experiments due to the time needed to make CT scans. Moreover, it leads to an inflation of the amount of data and thus to costly post-processing computations to quantify the dynamic behaviour from the sequence of time frames, hereby often ignoring the temporal correlations of the sample structure. Our proposed 4D $\mu$CT reconstruction technique, named DYRECT, estimates individual attenuation evolution profiles for each position in the sample. This leads to a novel memory-efficient event-based representation of the sample, using as little as three image volumes: its initial attenuation, its final attenuation and the transition times. This third volume represents local events on a continuous timescale instead of the discrete global time frames. We propose a method to iteratively reconstruct the transition times and the attenuation volumes. The dynamic reconstruction technique was validated on synthetic ground truth data and experimental data, and was found to effectively pinpoint the transition times in the synthetic dataset with a time resolution corresponding to less than a tenth of the amount of projections required to reconstruct traditional $\mu$CT time frames.
Inheriting Bayer's Legacy-Joint Remosaicing and Denoising for Quad Bayer Image Sensor
Mar 23, 2023Abstract:Pixel binning based Quad sensors have emerged as a promising solution to overcome the hardware limitations of compact cameras in low-light imaging. However, binning results in lower spatial resolution and non-Bayer CFA artifacts. To address these challenges, we propose a dual-head joint remosaicing and denoising network (DJRD), which enables the conversion of noisy Quad Bayer and standard noise-free Bayer pattern without any resolution loss. DJRD includes a newly designed Quad Bayer remosaicing (QB-Re) block, integrated denoising modules based on Swin-transformer and multi-scale wavelet transform. The QB-Re block constructs the convolution kernel based on the CFA pattern to achieve a periodic color distribution in the perceptual field, which is used to extract exact spectral information and reduce color misalignment. The integrated Swin-Transformer and multi-scale wavelet transform capture non-local dependencies, frequency and location information to effectively reduce practical noise. By identifying challenging patches utilizing Moire and zipper detection metrics, we enable our model to concentrate on difficult patches during the post-training phase, which enhances the model's performance in hard cases. Our proposed model outperforms competing models by approximately 3dB, without additional complexity in hardware or software.
Bayesian Deconvolution of Scanning Electron Microscopy Images Using Point-spread Function Estimation and Non-local Regularization
Oct 23, 2018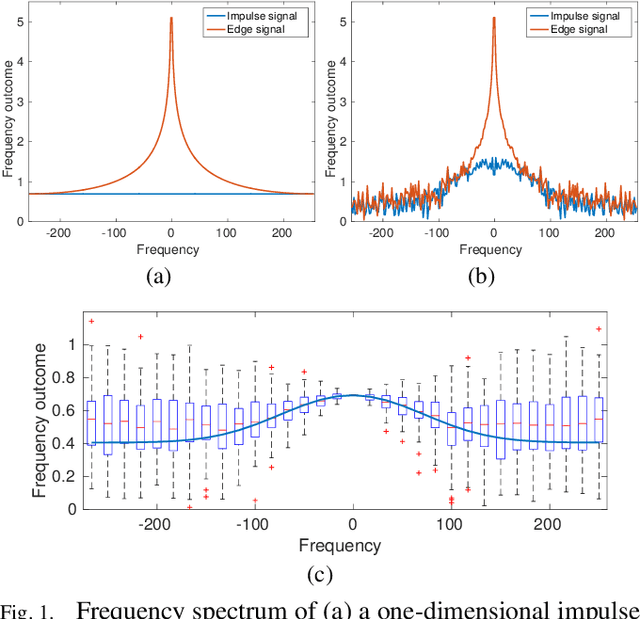
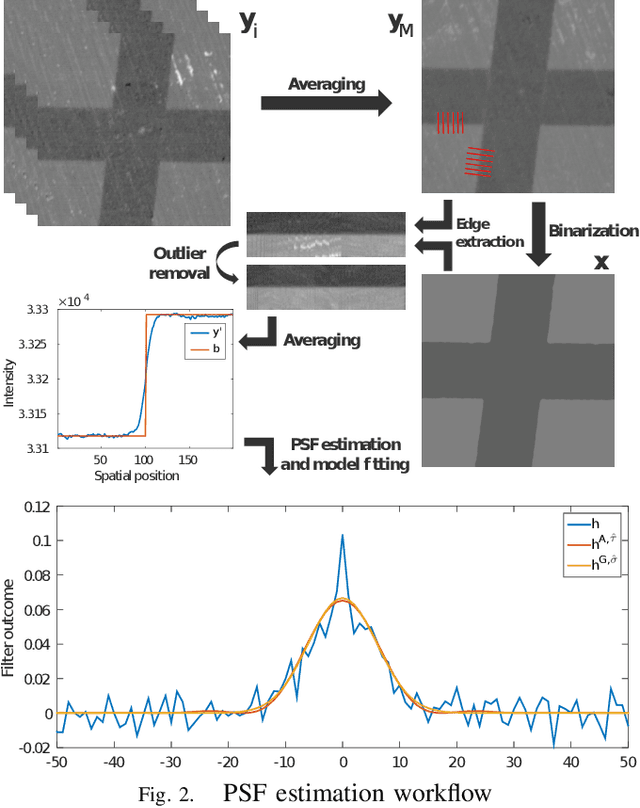
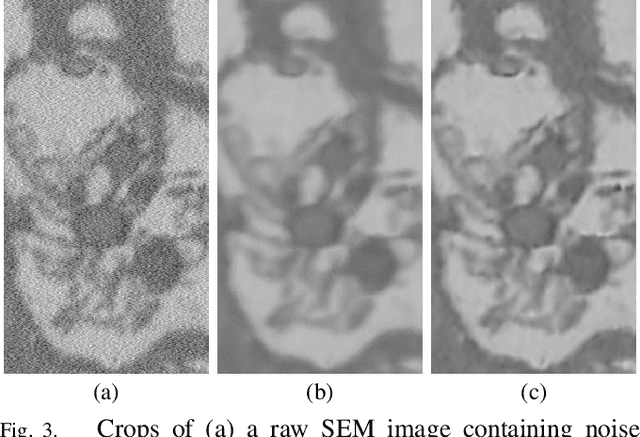
Abstract:Microscopy is one of the most essential imaging techniques in life sciences. High-quality images are required in order to solve (potentially life-saving) biomedical research problems. Many microscopy techniques do not achieve sufficient resolution for these purposes, being limited by physical diffraction and hardware deficiencies. Electron microscopy addresses optical diffraction by measuring emitted or transmitted electrons instead of photons, yielding nanometer resolution. Despite pushing back the diffraction limit, blur should still be taken into account because of practical hardware imperfections and remaining electron diffraction. Deconvolution algorithms can remove some of the blur in post-processing but they depend on knowledge of the point-spread function (PSF) and should accurately regularize noise. Any errors in the estimated PSF or noise model will reduce their effectiveness. This paper proposes a new procedure to estimate the lateral component of the point spread function of a 3D scanning electron microscope more accurately. We also propose a Bayesian maximum a posteriori deconvolution algorithm with a non-local image prior which employs this PSF estimate and previously developed noise statistics. We demonstrate visual quality improvements and show that applying our method improves the quality of subsequent segmentation steps.
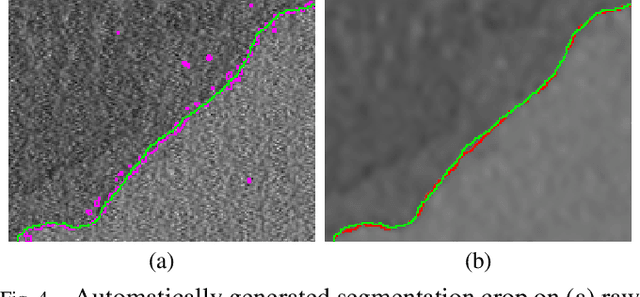
Convolutional Neural Network Pruning to Accelerate Membrane Segmentation in Electron Microscopy
Oct 23, 2018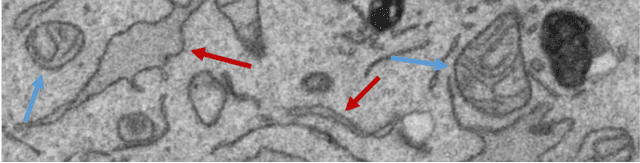
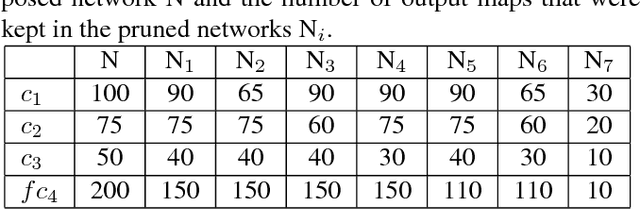
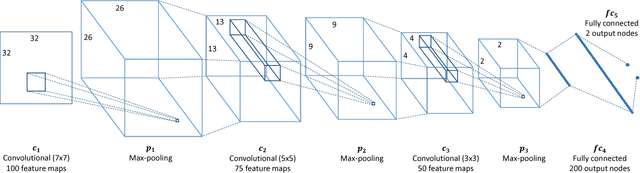
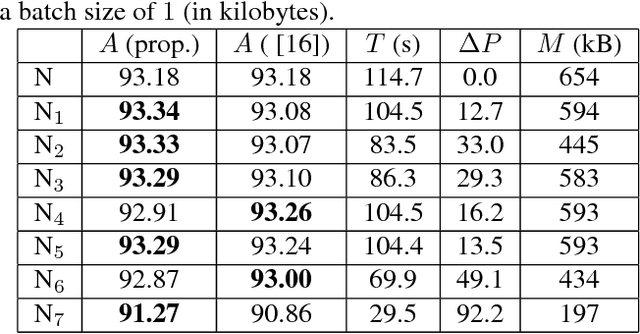
Abstract:Biological membranes are one of the most basic structures and regions of interest in cell biology. In the study of membranes, segment extraction is a well-known and difficult problem because of impeding noise, directional and thickness variability, etc. Recent advances in electron microscopy membrane segmentation are able to cope with such difficulties by training convolutional neural networks. However, because of the massive amount of features that have to be extracted while propagating forward, the practical usability diminishes, even with state-of-the-art GPU's. A significant part of these network features typically contains redundancy through correlation and sparsity. In this work, we propose a pruning method for convolutional neural networks that ensures the training loss increase is minimized. We show that the pruned networks, after retraining, are more efficient in terms of time and memory, without significantly affecting the network accuracy. This way, we manage to obtain real-time membrane segmentation performance, for our specific electron microscopy setup.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge