Jonas De Vylder
When normalization hallucinates: unseen risks in AI-powered whole slide image processing
Dec 08, 2025


Abstract:Whole slide image (WSI) normalization remains a vital preprocessing step in computational pathology. Increasingly driven by deep learning, these models learn to approximate data distributions from training examples. This often results in outputs that gravitate toward the average, potentially masking diagnostically important features. More critically, they can introduce hallucinated content, artifacts that appear realistic but are not present in the original tissue, posing a serious threat to downstream analysis. These hallucinations are nearly impossible to detect visually, and current evaluation practices often overlook them. In this work, we demonstrate that the risk of hallucinations is real and underappreciated. While many methods perform adequately on public datasets, we observe a concerning frequency of hallucinations when these same models are retrained and evaluated on real-world clinical data. To address this, we propose a novel image comparison measure designed to automatically detect hallucinations in normalized outputs. Using this measure, we systematically evaluate several well-cited normalization methods retrained on real-world data, revealing significant inconsistencies and failures that are not captured by conventional metrics. Our findings underscore the need for more robust, interpretable normalization techniques and stricter validation protocols in clinical deployment.
Bayesian Deconvolution of Scanning Electron Microscopy Images Using Point-spread Function Estimation and Non-local Regularization
Oct 23, 2018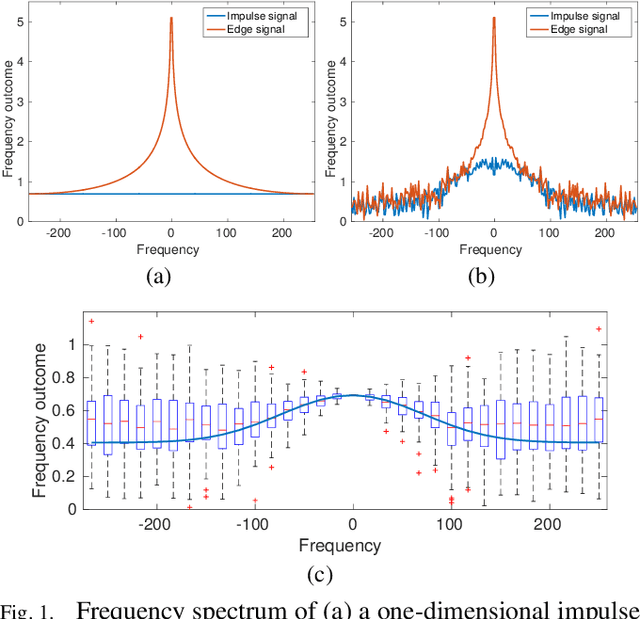
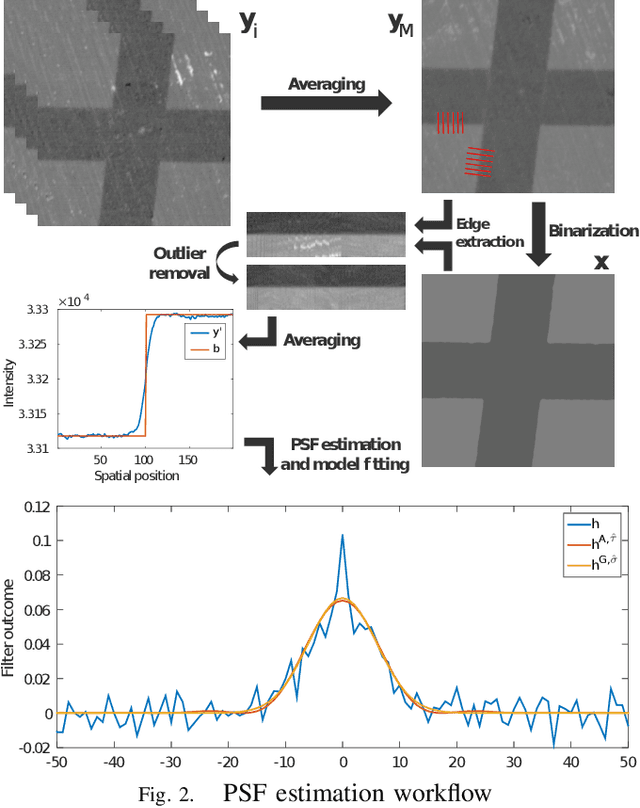
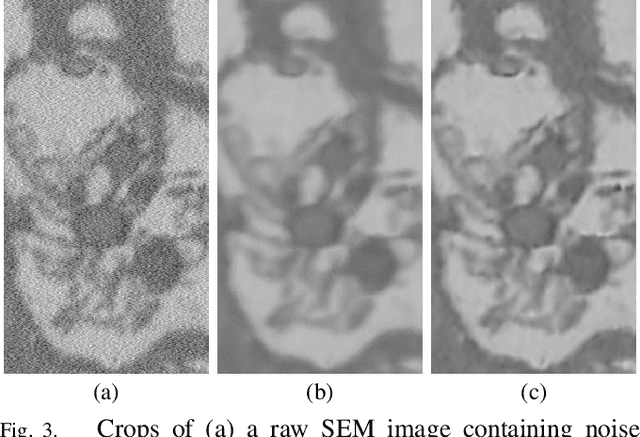
Abstract:Microscopy is one of the most essential imaging techniques in life sciences. High-quality images are required in order to solve (potentially life-saving) biomedical research problems. Many microscopy techniques do not achieve sufficient resolution for these purposes, being limited by physical diffraction and hardware deficiencies. Electron microscopy addresses optical diffraction by measuring emitted or transmitted electrons instead of photons, yielding nanometer resolution. Despite pushing back the diffraction limit, blur should still be taken into account because of practical hardware imperfections and remaining electron diffraction. Deconvolution algorithms can remove some of the blur in post-processing but they depend on knowledge of the point-spread function (PSF) and should accurately regularize noise. Any errors in the estimated PSF or noise model will reduce their effectiveness. This paper proposes a new procedure to estimate the lateral component of the point spread function of a 3D scanning electron microscope more accurately. We also propose a Bayesian maximum a posteriori deconvolution algorithm with a non-local image prior which employs this PSF estimate and previously developed noise statistics. We demonstrate visual quality improvements and show that applying our method improves the quality of subsequent segmentation steps.
Convolutional Neural Network Pruning to Accelerate Membrane Segmentation in Electron Microscopy
Oct 23, 2018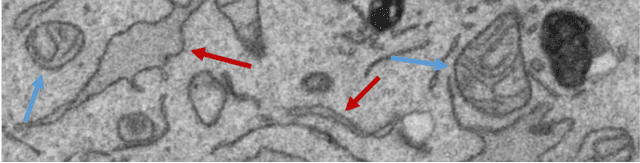
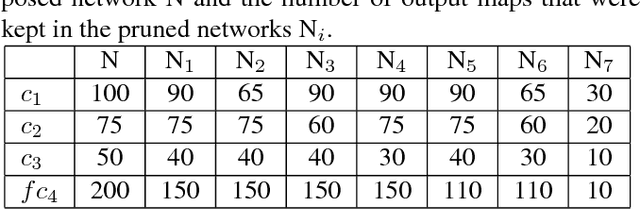
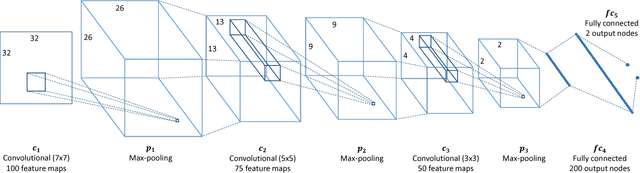
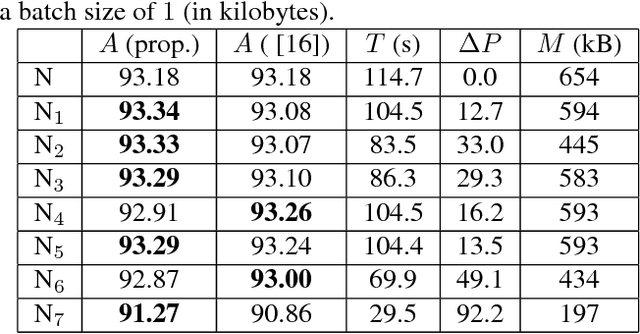
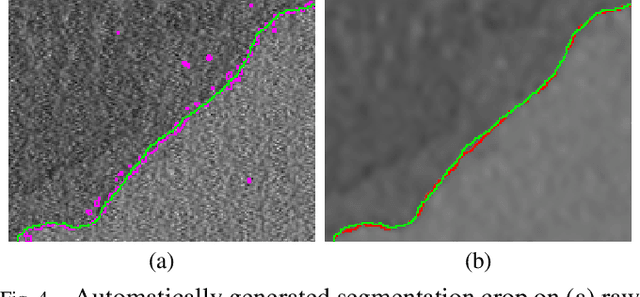
Abstract:Biological membranes are one of the most basic structures and regions of interest in cell biology. In the study of membranes, segment extraction is a well-known and difficult problem because of impeding noise, directional and thickness variability, etc. Recent advances in electron microscopy membrane segmentation are able to cope with such difficulties by training convolutional neural networks. However, because of the massive amount of features that have to be extracted while propagating forward, the practical usability diminishes, even with state-of-the-art GPU's. A significant part of these network features typically contains redundancy through correlation and sparsity. In this work, we propose a pruning method for convolutional neural networks that ensures the training loss increase is minimized. We show that the pruned networks, after retraining, are more efficient in terms of time and memory, without significantly affecting the network accuracy. This way, we manage to obtain real-time membrane segmentation performance, for our specific electron microscopy setup.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge