Bayesian Deconvolution of Scanning Electron Microscopy Images Using Point-spread Function Estimation and Non-local Regularization
Paper and Code
Oct 23, 2018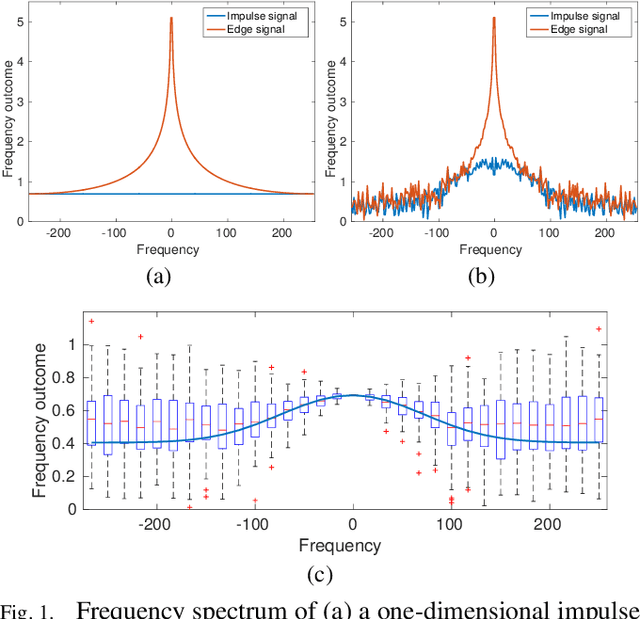
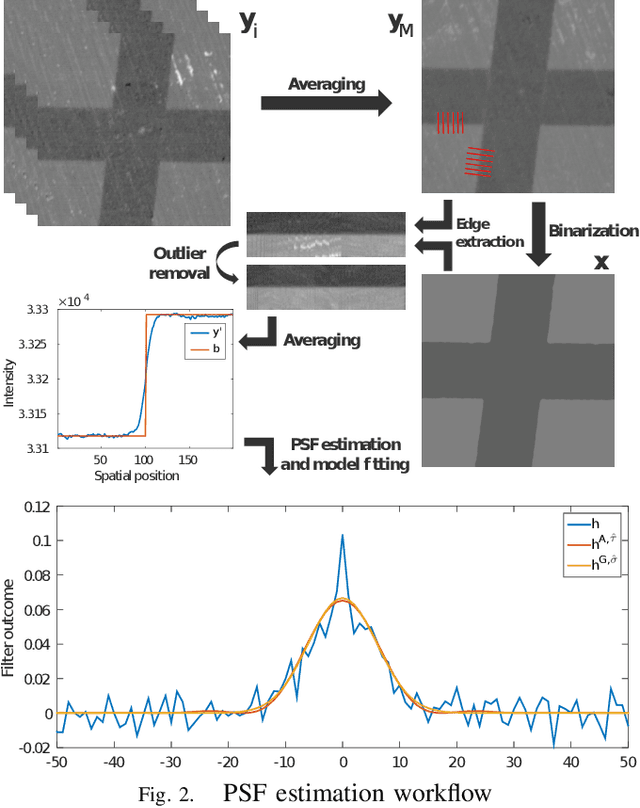
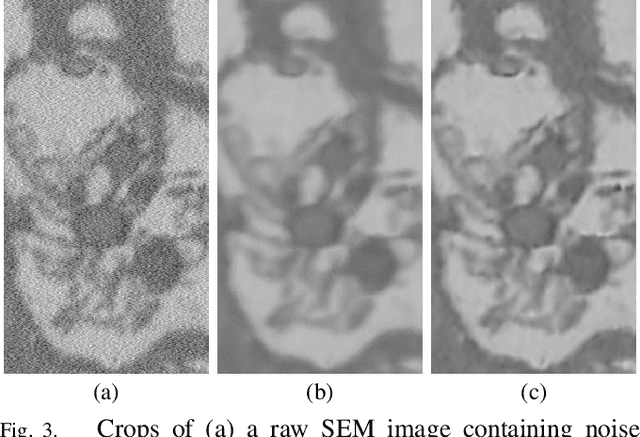
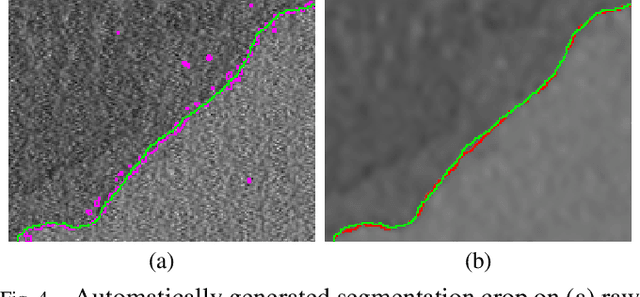
Microscopy is one of the most essential imaging techniques in life sciences. High-quality images are required in order to solve (potentially life-saving) biomedical research problems. Many microscopy techniques do not achieve sufficient resolution for these purposes, being limited by physical diffraction and hardware deficiencies. Electron microscopy addresses optical diffraction by measuring emitted or transmitted electrons instead of photons, yielding nanometer resolution. Despite pushing back the diffraction limit, blur should still be taken into account because of practical hardware imperfections and remaining electron diffraction. Deconvolution algorithms can remove some of the blur in post-processing but they depend on knowledge of the point-spread function (PSF) and should accurately regularize noise. Any errors in the estimated PSF or noise model will reduce their effectiveness. This paper proposes a new procedure to estimate the lateral component of the point spread function of a 3D scanning electron microscope more accurately. We also propose a Bayesian maximum a posteriori deconvolution algorithm with a non-local image prior which employs this PSF estimate and previously developed noise statistics. We demonstrate visual quality improvements and show that applying our method improves the quality of subsequent segmentation steps.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge