Islam Akef Ebeid
Inferred global dense residue transition graphs from primary structure sequences enable protein interaction prediction via directed graph convolutional neural networks
Oct 15, 2025Abstract:Introduction Accurate prediction of protein-protein interactions (PPIs) is crucial for understanding cellular functions and advancing drug development. Existing in-silico methods use direct sequence embeddings from Protein Language Models (PLMs). Others use Graph Neural Networks (GNNs) for 3D protein structures. This study explores less computationally intensive alternatives. We introduce a novel framework for downstream PPI prediction through link prediction. Methods We introduce a two-stage graph representation learning framework, ProtGram-DirectGCN. First, we developed ProtGram. This approach models a protein's primary structure as a hierarchy of globally inferred n-gram graphs. In these graphs, residue transition probabilities define edge weights. Each edge connects a pair of residues in a directed graph. The probabilities are aggregated from a large corpus of sequences. Second, we propose DirectGCN, a custom directed graph convolutional neural network. This model features a unique convolutional layer. It processes information through separate path-specific transformations: incoming, outgoing, and undirected. A shared transformation is also applied. These paths are combined via a learnable gating mechanism. We apply DirectGCN to ProtGram graphs to learn residue-level embeddings. These embeddings are pooled via attention to generate protein-level embeddings for prediction. Results We first established the efficacy of DirectGCN on standard node classification benchmarks. Its performance matches established methods on general datasets. The model excels at complex, directed graphs with dense, heterophilic structures. When applied to PPI prediction, the full ProtGram-DirectGCN framework delivers robust predictive power. This strong performance holds even with limited training data.
MedGraph: An experimental semantic information retrieval method using knowledge graph embedding for the biomedical citations indexed in PubMed
Dec 14, 2021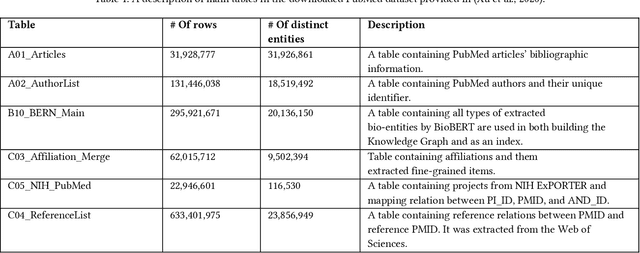
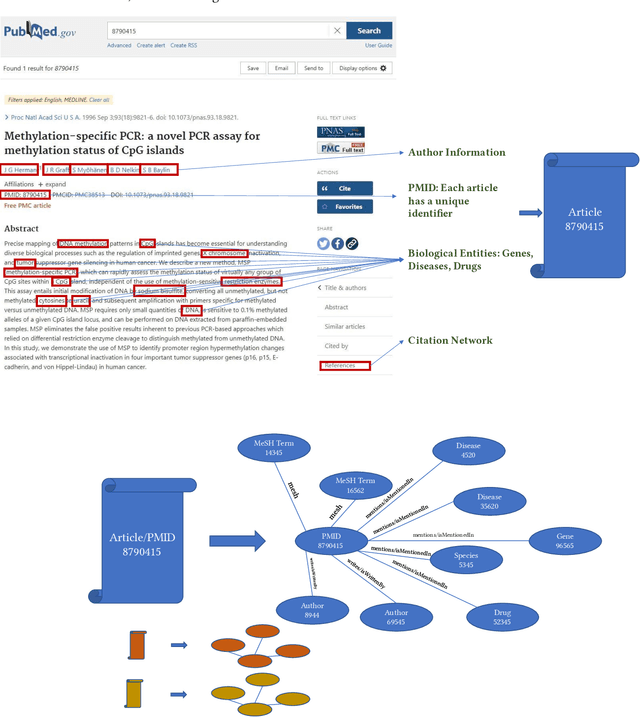
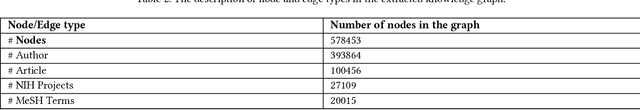
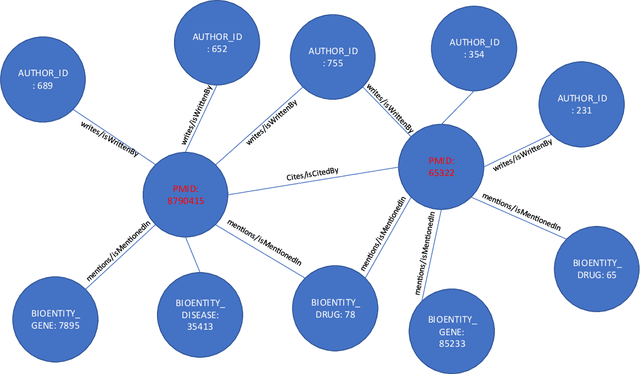
Abstract:Here we study the semantic search and retrieval problem in biomedical digital libraries. First, we introduce MedGraph, a knowledge graph embedding-based method that provides semantic relevance retrieval and ranking for the biomedical literature indexed in PubMed. Second, we evaluate our method using PubMed's Best Match algorithm. Moreover, we compare our method MedGraph to a traditional TFIDF based algorithm. We use a dataset extracted from PubMed, including 30 million articles' metadata such as abstracts, author information, citation information, and extracted biological entity mentions. We do that by pulling a subset of the dataset to evaluate MedGraph using predefined queries with ground truth ranked results. To our knowledge, this technique has not been explored before in biomedical information retrieval. In addition, our results provide evidence that semantic approaches to search and relevance in biomedical digital libraries that rely on knowledge graph modeling offer better search relevance results when compared with traditional approaches in terms of objective metrics.
Graph-based hierarchical record clustering for unsupervised entity resolution
Dec 12, 2021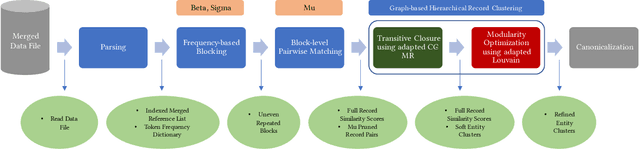

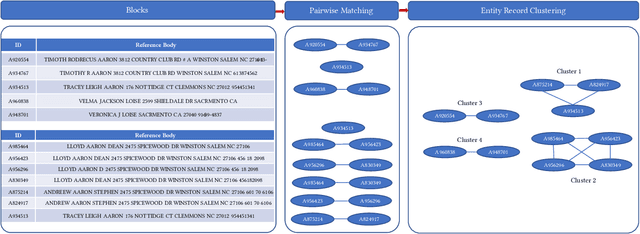
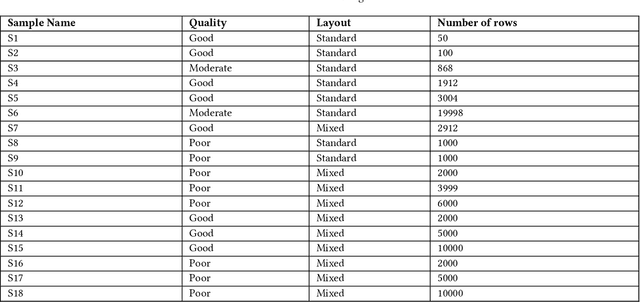
Abstract:Here we study the problem of matched record clustering in unsupervised entity resolution. We build upon a state-of-the-art probabilistic framework named the Data Washing Machine (DWM). We introduce a graph-based hierarchical 2-step record clustering method (GDWM) that first identifies large, connected components or, as we call them, soft clusters in the matched record pairs using a graph-based transitive closure algorithm utilized in the DWM. That is followed by breaking down the discovered soft clusters into more precise entity clusters in a hierarchical manner using an adapted graph-based modularity optimization method. Our approach provides several advantages over the original implementation of the DWM, mainly a significant speed-up, increased precision, and overall increased F1 scores. We demonstrate the efficacy of our approach using experiments on multiple synthetic datasets. Our results also provide evidence of the utility of graph theory-based algorithms despite their sparsity in the literature on unsupervised entity resolution.
Biomedical Knowledge Graph Refinement and Completion using Graph Representation Learning and Top-K Similarity Measure
Dec 18, 2020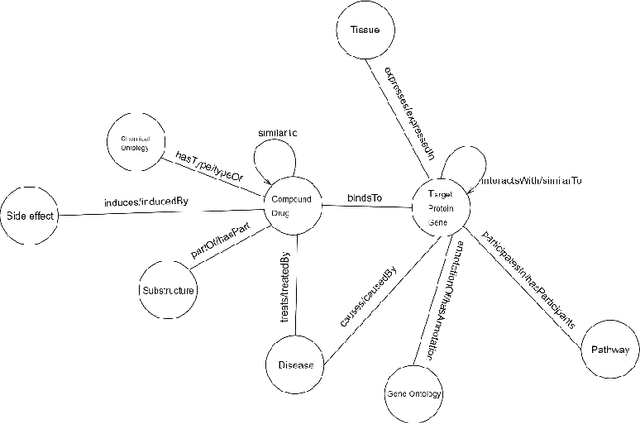

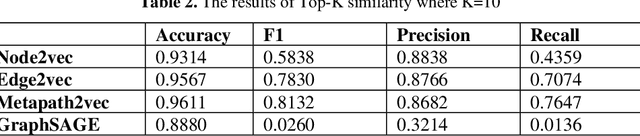
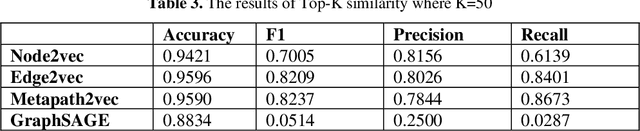
Abstract:Knowledge Graphs have been one of the fundamental methods for integrating heterogeneous data sources. Integrating heterogeneous data sources is crucial, especially in the biomedical domain, where central data-driven tasks such as drug discovery rely on incorporating information from different biomedical databases. These databases contain various biological entities and relations such as proteins (PDB), genes (Gene Ontology), drugs (DrugBank), diseases (DDB), and protein-protein interactions (BioGRID). The process of semantically integrating heterogeneous biomedical databases is often riddled with imperfections. The quality of data-driven drug discovery relies on the accuracy of the mining methods used and the data's quality as well. Thus, having complete and refined biomedical knowledge graphs is central to achieving more accurate drug discovery outcomes. Here we propose using the latest graph representation learning and embedding models to refine and complete biomedical knowledge graphs. This preliminary work demonstrates learning discrete representations of the integrated biomedical knowledge graph Chem2Bio2RD [3]. We perform a knowledge graph completion and refinement task using a simple top-K cosine similarity measure between the learned embedding vectors to predict missing links between drugs and targets present in the data. We show that this simple procedure can be used alternatively to binary classifiers in link prediction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge