MedGraph: An experimental semantic information retrieval method using knowledge graph embedding for the biomedical citations indexed in PubMed
Paper and Code
Dec 14, 2021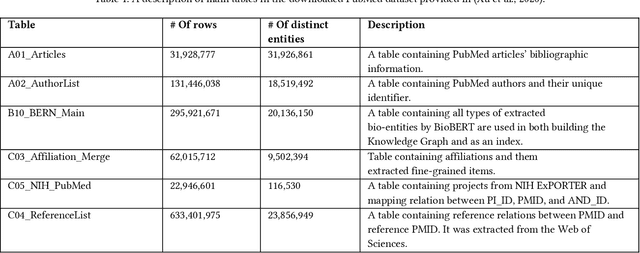
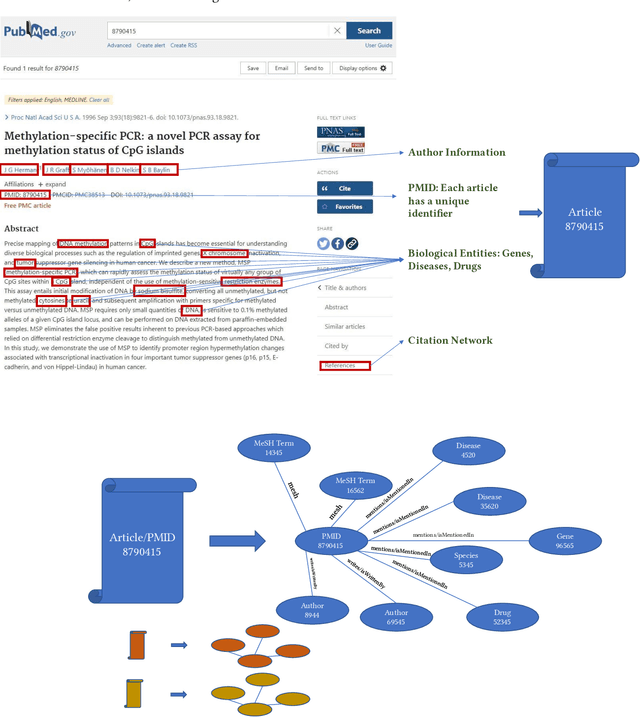
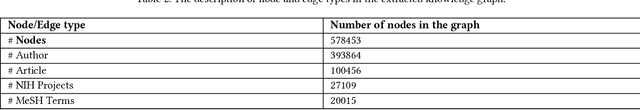
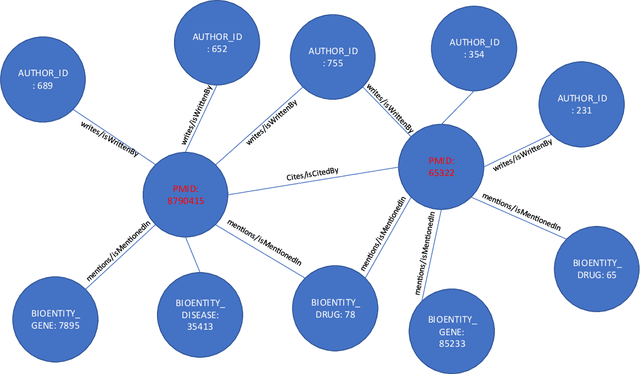
Here we study the semantic search and retrieval problem in biomedical digital libraries. First, we introduce MedGraph, a knowledge graph embedding-based method that provides semantic relevance retrieval and ranking for the biomedical literature indexed in PubMed. Second, we evaluate our method using PubMed's Best Match algorithm. Moreover, we compare our method MedGraph to a traditional TFIDF based algorithm. We use a dataset extracted from PubMed, including 30 million articles' metadata such as abstracts, author information, citation information, and extracted biological entity mentions. We do that by pulling a subset of the dataset to evaluate MedGraph using predefined queries with ground truth ranked results. To our knowledge, this technique has not been explored before in biomedical information retrieval. In addition, our results provide evidence that semantic approaches to search and relevance in biomedical digital libraries that rely on knowledge graph modeling offer better search relevance results when compared with traditional approaches in terms of objective metrics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge