Jack Roper
Transformer-based Program Synthesis for Low-Data Environments
May 18, 2022


Abstract:Recent advancements in large pre-trained transformer models (GPT2/3, T5) have found use in program synthesis to generate programs that satisfy a set of input/output examples. However, these models perform poorly on long-horizon and low-data tasks, and often don't seem to understand the semantics of the languages they generate. We investigate an approach that tackles both of these issues, by using attributed context-free-grammars of programming languages to generate programs, and then analyzing generated programs so that they can be annotated with compile and runtime attributes, such as types, so that information about the program can be remembered during long-horizon generation. We firstly find that synthesized datasets can be made efficiently and can provide transformer models with enough data in order to perform well on some synthesis tasks. We also find that giving models access to program attributes is especially effective in low-data environments, and tends improve the quality and reduce errors of transformer-generated programs.
Biomedical Knowledge Graph Refinement and Completion using Graph Representation Learning and Top-K Similarity Measure
Dec 18, 2020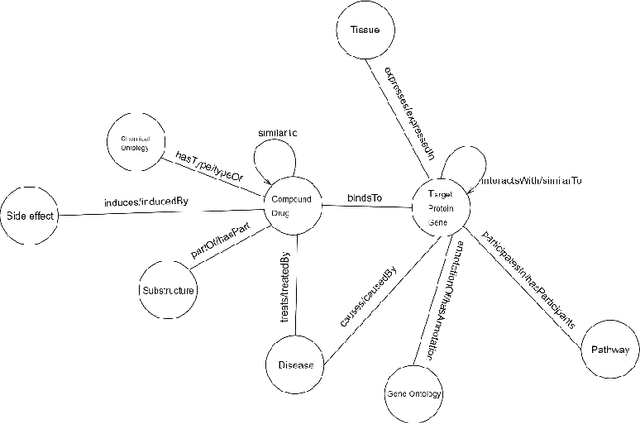

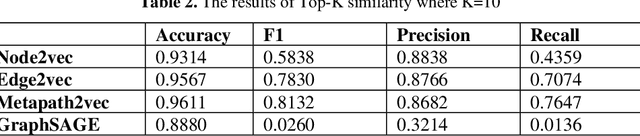
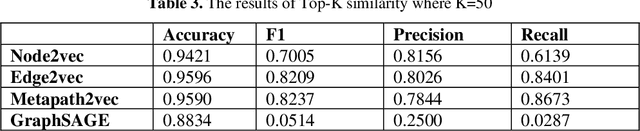
Abstract:Knowledge Graphs have been one of the fundamental methods for integrating heterogeneous data sources. Integrating heterogeneous data sources is crucial, especially in the biomedical domain, where central data-driven tasks such as drug discovery rely on incorporating information from different biomedical databases. These databases contain various biological entities and relations such as proteins (PDB), genes (Gene Ontology), drugs (DrugBank), diseases (DDB), and protein-protein interactions (BioGRID). The process of semantically integrating heterogeneous biomedical databases is often riddled with imperfections. The quality of data-driven drug discovery relies on the accuracy of the mining methods used and the data's quality as well. Thus, having complete and refined biomedical knowledge graphs is central to achieving more accurate drug discovery outcomes. Here we propose using the latest graph representation learning and embedding models to refine and complete biomedical knowledge graphs. This preliminary work demonstrates learning discrete representations of the integrated biomedical knowledge graph Chem2Bio2RD [3]. We perform a knowledge graph completion and refinement task using a simple top-K cosine similarity measure between the learned embedding vectors to predict missing links between drugs and targets present in the data. We show that this simple procedure can be used alternatively to binary classifiers in link prediction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge