Ilija Tavchioski
Detection of depression on social networks using transformers and ensembles
May 09, 2023

Abstract:As the impact of technology on our lives is increasing, we witness increased use of social media that became an essential tool not only for communication but also for sharing information with community about our thoughts and feelings. This can be observed also for people with mental health disorders such as depression where they use social media for expressing their thoughts and asking for help. This opens a possibility to automatically process social media posts and detect signs of depression. We build several large pre-trained language model based classifiers for depression detection from social media posts. Besides fine-tuning BERT, RoBERTA, BERTweet, and mentalBERT were also construct two types of ensembles. We analyze the performance of our models on two data sets of posts from social platforms Reddit and Twitter, and investigate also the performance of transfer learning across the two data sets. The results show that transformer ensembles improve over the single transformer-based classifiers.
Multi-label classification for biomedical literature: an overview of the BioCreative VII LitCovid Track for COVID-19 literature topic annotations
Apr 20, 2022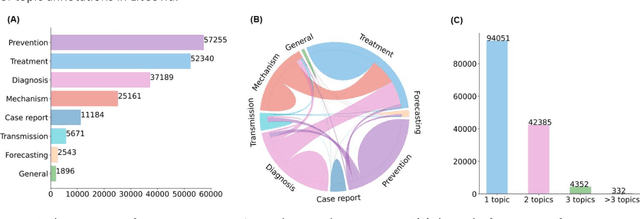
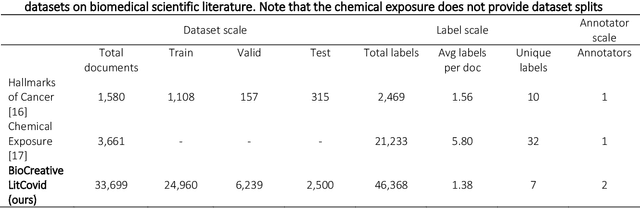
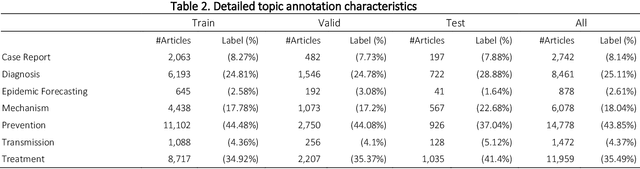
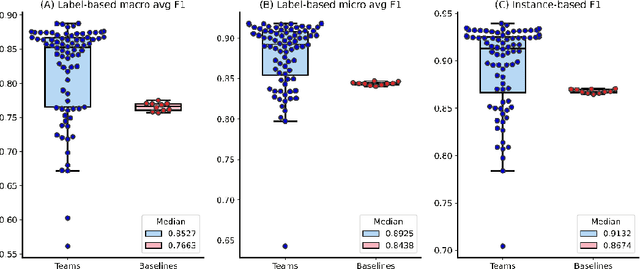
Abstract:The COVID-19 pandemic has been severely impacting global society since December 2019. Massive research has been undertaken to understand the characteristics of the virus and design vaccines and drugs. The related findings have been reported in biomedical literature at a rate of about 10,000 articles on COVID-19 per month. Such rapid growth significantly challenges manual curation and interpretation. For instance, LitCovid is a literature database of COVID-19-related articles in PubMed, which has accumulated more than 200,000 articles with millions of accesses each month by users worldwide. One primary curation task is to assign up to eight topics (e.g., Diagnosis and Treatment) to the articles in LitCovid. Despite the continuing advances in biomedical text mining methods, few have been dedicated to topic annotations in COVID-19 literature. To close the gap, we organized the BioCreative LitCovid track to call for a community effort to tackle automated topic annotation for COVID-19 literature. The BioCreative LitCovid dataset, consisting of over 30,000 articles with manually reviewed topics, was created for training and testing. It is one of the largest multilabel classification datasets in biomedical scientific literature. 19 teams worldwide participated and made 80 submissions in total. Most teams used hybrid systems based on transformers. The highest performing submissions achieved 0.8875, 0.9181, and 0.9394 for macro F1-score, micro F1-score, and instance-based F1-score, respectively. The level of participation and results demonstrate a successful track and help close the gap between dataset curation and method development. The dataset is publicly available via https://ftp.ncbi.nlm.nih.gov/pub/lu/LitCovid/biocreative/ for benchmarking and further development.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge