Hwihun Jeong
Efficient and robust 3D blind harmonization for large domain gaps
Apr 30, 2025Abstract:Blind harmonization has emerged as a promising technique for MR image harmonization to achieve scale-invariant representations, requiring only target domain data (i.e., no source domain data necessary). However, existing methods face limitations such as inter-slice heterogeneity in 3D, moderate image quality, and limited performance for a large domain gap. To address these challenges, we introduce BlindHarmonyDiff, a novel blind 3D harmonization framework that leverages an edge-to-image model tailored specifically to harmonization. Our framework employs a 3D rectified flow trained on target domain images to reconstruct the original image from an edge map, then yielding a harmonized image from the edge of a source domain image. We propose multi-stride patch training for efficient 3D training and a refinement module for robust inference by suppressing hallucination. Extensive experiments demonstrate that BlindHarmonyDiff outperforms prior arts by harmonizing diverse source domain images to the target domain, achieving higher correspondence to the target domain characteristics. Downstream task-based quality assessments such as tissue segmentation and age prediction on diverse MR scanners further confirm the effectiveness of our approach and demonstrate the capability of our robust and generalizable blind harmonization.
$χ$-sepnet: Deep neural network for magnetic susceptibility source separation
Sep 24, 2024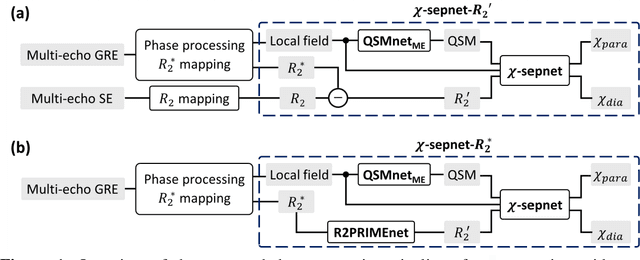
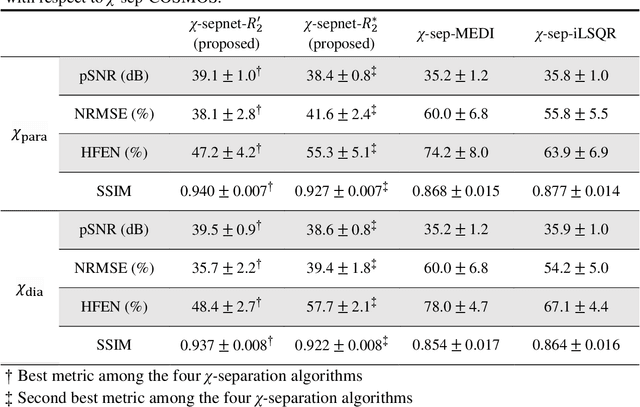
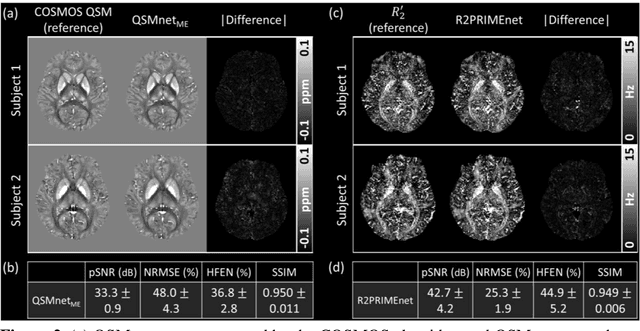
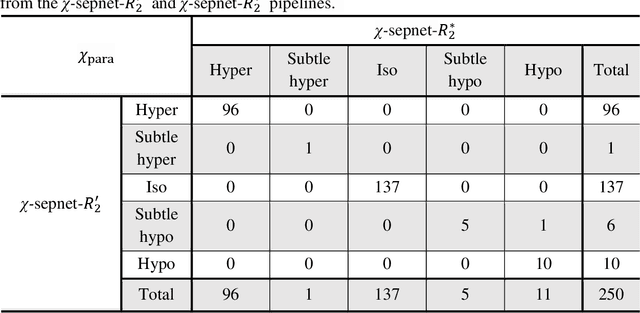
Abstract:Magnetic susceptibility source separation ($\chi$-separation), an advanced quantitative susceptibility mapping (QSM) method, enables the separate estimation of para- and diamagnetic susceptibility source distributions in the brain. The method utilizes reversible transverse relaxation (R2'=R2*-R2) to complement frequency shift information for estimating susceptibility source concentrations, requiring time-consuming data acquisition for R2 in addition R2*. To address this challenge, we develop a new deep learning network, $\chi$-sepnet, and propose two deep learning-based susceptibility source separation pipelines, $\chi$-sepnet-R2' for inputs with multi-echo GRE and multi-echo spin-echo, and $\chi$-sepnet-R2* for input with multi-echo GRE only. $\chi$-sepnet is trained using multiple head orientation data that provide streaking artifact-free labels, generating high-quality $\chi$-separation maps. The evaluation of the pipelines encompasses both qualitative and quantitative assessments in healthy subjects, and visual inspection of lesion characteristics in multiple sclerosis patients. The susceptibility source-separated maps of the proposed pipelines delineate detailed brain structures with substantially reduced artifacts compared to those from conventional regularization-based reconstruction methods. In quantitative analysis, $\chi$-sepnet-R2' achieves the best outcomes followed by $\chi$-sepnet-R2*, outperforming the conventional methods. When the lesions of multiple sclerosis patients are assessed, both pipelines report identical lesion characteristics in most lesions ($\chi$para: 99.6% and $\chi$dia: 98.4% out of 250 lesions). The $\chi$-sepnet-R2* pipeline, which only requires multi-echo GRE data, has demonstrated its potential to offer broad clinical and scientific applications, although further evaluations for various diseases and pathological conditions are necessary.
MOST: MR reconstruction Optimization for multiple downStream Tasks via continual learning
Sep 16, 2024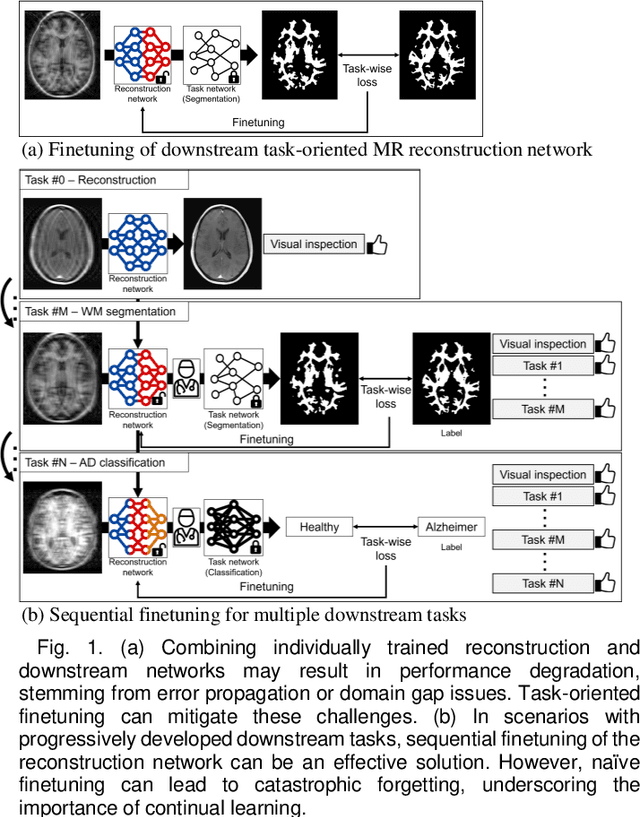

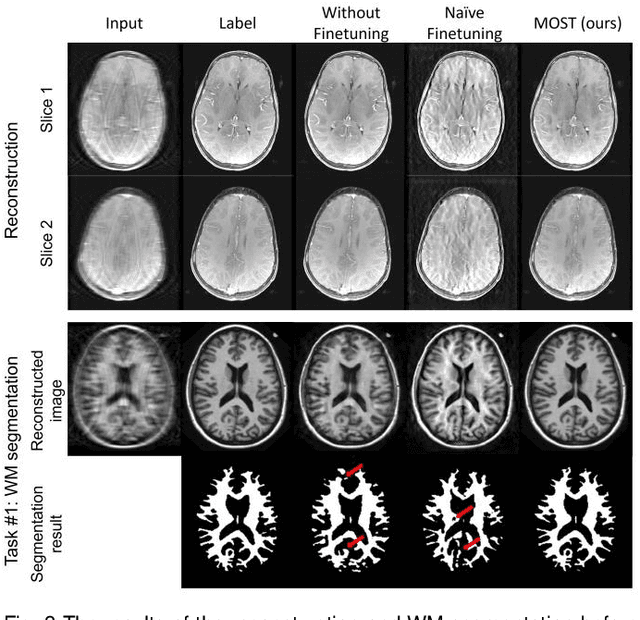
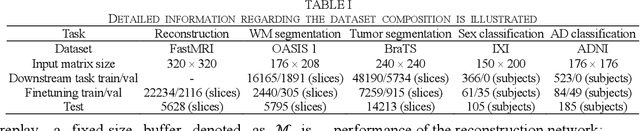
Abstract:Deep learning-based Magnetic Resonance (MR) reconstruction methods have focused on generating high-quality images but they often overlook the impact on downstream tasks (e.g., segmentation) that utilize the reconstructed images. Cascading separately trained reconstruction network and downstream task network has been shown to introduce performance degradation due to error propagation and domain gaps between training datasets. To mitigate this issue, downstream task-oriented reconstruction optimization has been proposed for a single downstream task. Expanding this optimization to multi-task scenarios is not straightforward. In this work, we extended this optimization to sequentially introduced multiple downstream tasks and demonstrated that a single MR reconstruction network can be optimized for multiple downstream tasks by deploying continual learning (MOST). MOST integrated techniques from replay-based continual learning and image-guided loss to overcome catastrophic forgetting. Comparative experiments demonstrated that MOST outperformed a reconstruction network without finetuning, a reconstruction network with na\"ive finetuning, and conventional continual learning methods. This advancement empowers the application of a single MR reconstruction network for multiple downstream tasks. The source code is available at: https://github.com/SNU-LIST/MOST
BlindHarmony: "Blind" Harmonization for MR Images via Flow model
May 18, 2023Abstract:In MRI, images of the same contrast (e.g., T1) from the same subject can show noticeable differences when acquired using different hardware, sequences, or scan parameters. These differences in images create a domain gap that needs to be bridged by a step called image harmonization, in order to process the images successfully using conventional or deep learning-based image analysis (e.g., segmentation). Several methods, including deep learning-based approaches, have been proposed to achieve image harmonization. However, they often require datasets of multiple characteristics for deep learning training and may still be unsuccessful when applied to images of an unseen domain. To address this limitation, we propose a novel concept called "Blind Harmonization," which utilizes only target domain data for training but still has the capability of harmonizing unseen domain images. For the implementation of Blind Harmonization, we developed BlindHarmony using an unconditional flow model trained on target domain data. The harmonized image is optimized to have a correlation with the input source domain image while ensuring that the latent vector of the flow model is close to the center of the Gaussian. BlindHarmony was evaluated using simulated and real datasets and compared with conventional methods. BlindHarmony achieved a noticeable performance in both datasets, highlighting its potential for future use in clinical settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge