Hui Che
Residual Dense Swin Transformer for Continuous Depth-Independent Ultrasound Imaging
Mar 25, 2024Abstract:Ultrasound imaging is crucial for evaluating organ morphology and function, yet depth adjustment can degrade image quality and field-of-view, presenting a depth-dependent dilemma. Traditional interpolation-based zoom-in techniques often sacrifice detail and introduce artifacts. Motivated by the potential of arbitrary-scale super-resolution to naturally address these inherent challenges, we present the Residual Dense Swin Transformer Network (RDSTN), designed to capture the non-local characteristics and long-range dependencies intrinsic to ultrasound images. It comprises a linear embedding module for feature enhancement, an encoder with shifted-window attention for modeling non-locality, and an MLP decoder for continuous detail reconstruction. This strategy streamlines balancing image quality and field-of-view, which offers superior textures over traditional methods. Experimentally, RDSTN outperforms existing approaches while requiring fewer parameters. In conclusion, RDSTN shows promising potential for ultrasound image enhancement by overcoming the limitations of conventional interpolation-based methods and achieving depth-independent imaging.
* Accepted by ICASSP2024, https://ieeexplore.ieee.org/document/10447712
Colorectal Polyp Classification from White-light Colonoscopy Images via Domain Alignment
Aug 05, 2021



Abstract:Differentiation of colorectal polyps is an important clinical examination. A computer-aided diagnosis system is required to assist accurate diagnosis from colonoscopy images. Most previous studies at-tempt to develop models for polyp differentiation using Narrow-Band Imaging (NBI) or other enhanced images. However, the wide range of these models' applications for clinical work has been limited by the lagging of imaging techniques. Thus, we propose a novel framework based on a teacher-student architecture for the accurate colorectal polyp classification (CPC) through directly using white-light (WL) colonoscopy images in the examination. In practice, during training, the auxiliary NBI images are utilized to train a teacher network and guide the student network to acquire richer feature representation from WL images. The feature transfer is realized by domain alignment and contrastive learning. Eventually the final student network has the ability to extract aligned features from only WL images to facilitate the CPC task. Besides, we release the first public-available paired CPC dataset containing WL-NBI pairs for the alignment training. Quantitative and qualitative evaluation indicates that the proposed method outperforms the previous methods in CPC, improving the accuracy by 5.6%with very fast speed.
Realistic Ultrasound Image Synthesis for Improved Classification of Liver Disease
Jul 27, 2021



Abstract:With the success of deep learning-based methods applied in medical image analysis, convolutional neural networks (CNNs) have been investigated for classifying liver disease from ultrasound (US) data. However, the scarcity of available large-scale labeled US data has hindered the success of CNNs for classifying liver disease from US data. In this work, we propose a novel generative adversarial network (GAN) architecture for realistic diseased and healthy liver US image synthesis. We adopt the concept of stacking to synthesize realistic liver US data. Quantitative and qualitative evaluation is performed on 550 in-vivo B-mode liver US images collected from 55 subjects. We also show that the synthesized images, together with real in vivo data, can be used to significantly improve the performance of traditional CNN architectures for Nonalcoholic fatty liver disease (NAFLD) classification.
Multi-Feature Multi-Scale CNN-Derived COVID-19 Classification from Lung Ultrasound Data
Feb 23, 2021


Abstract:The global pandemic of the novel coronavirus disease 2019 (COVID-19) has put tremendous pressure on the medical system. Imaging plays a complementary role in the management of patients with COVID-19. Computed tomography (CT) and chest X-ray (CXR) are the two dominant screening tools. However, difficulty in eliminating the risk of disease transmission, radiation exposure and not being costeffective are some of the challenges for CT and CXR imaging. This fact induces the implementation of lung ultrasound (LUS) for evaluating COVID-19 due to its practical advantages of noninvasiveness, repeatability, and sensitive bedside property. In this paper, we utilize a deep learning model to perform the classification of COVID-19 from LUS data, which could produce objective diagnostic information for clinicians. Specifically, all LUS images are processed to obtain their corresponding local phase filtered images and radial symmetry transformed images before fed into the multi-scale residual convolutional neural network (CNN). Secondly, image combination as the input of the network is used to explore rich and reliable features. Feature fusion strategy at different levels is adopted to investigate the relationship between the depth of feature aggregation and the classification accuracy. Our proposed method is evaluated on the point-of-care US (POCUS) dataset together with the Italian COVID-19 Lung US database (ICLUS-DB) and shows promising performance for COVID-19 prediction.
Probability Map Guided Bi-directional Recurrent UNet for Pancreas Segmentation
Apr 07, 2019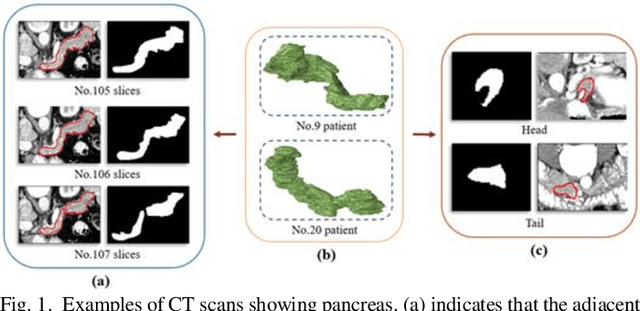
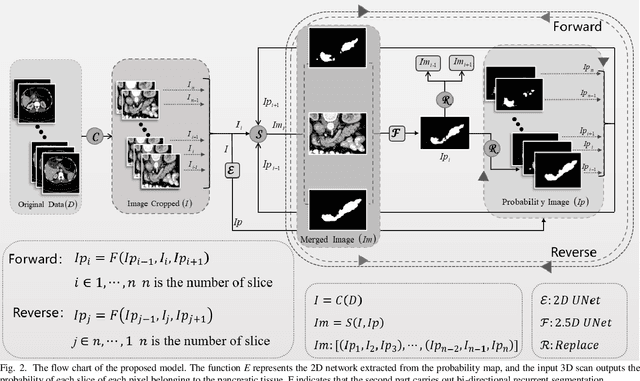
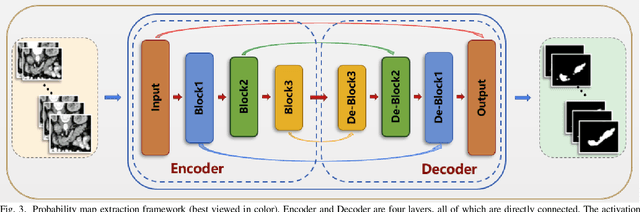
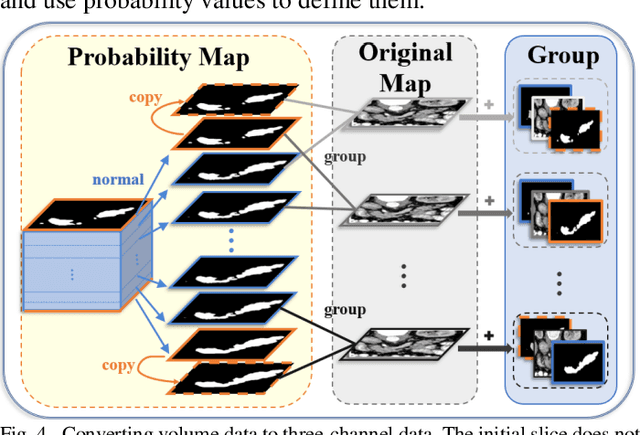
Abstract:Pancreatic cancer is one of the most lethal cancers as morbidity approximates mortality. A method for accurately segmenting the pancreas can assist doctors in the diagnosis and treatment of pancreatic cancer, but huge differences in shape and volume bring difficulties in segmentation. Among the current widely used approaches, the 2D method ignores the spatial information, and the 3D model is limited by high resource consumption and GPU memory occupancy. To address these issues, we propose a bi-directional recurrent UNet based on probabilistic map guidance (PBR-UNet). PBR-UNet includes a feature extraction module for extracting pixel-level probabilistic maps and a bi-directional recurrent module for fine segmentation. The extracted probabilistic map will be used to guide the fine segmentation and bi-directional recurrent module integrates contextual information into the entire network to avoid the loss of spatial information in propagation. By combining the probabilistic map of the adjacent slices with the bi-directional recurrent segmentation of intermediary slice, this paper solves the problem that the 2D network loses three-dimensional information and the 3D model leads to large computational resource consumption. We used Dice similarity coefficients (DSC) to evaluate our approach on NIH pancreatic datasets and eventually achieved a competitive result of 83.35%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge