Hugo Soulat
Unsupervised representational learning with recognition-parametrised probabilistic models
Sep 13, 2022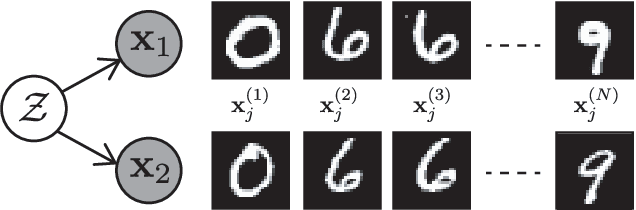
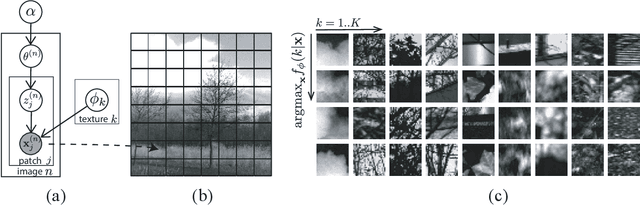
Abstract:We introduce a new approach to probabilistic unsupervised learning based on the recognition-parametrised model (RPM): a normalised semi-parametric hypothesis class for joint distributions over observed and latent variables. Under the key assumption that observations are conditionally independent given the latents, RPMs directly encode the "recognition" process, parametrising both the prior distribution on the latents and their conditional distributions given observations. This recognition model is paired with non-parametric descriptions of the marginal distribution of each observed variable. Thus, the focus is on learning a good latent representation that captures dependence between the measurements. The RPM permits exact maximum likelihood learning in settings with discrete latents and a tractable prior, even when the mapping between continuous observations and the latents is expressed through a flexible model such as a neural network. We develop effective approximations for the case of continuous latent variables with tractable priors. Unlike the approximations necessary in dual-parametrised models such as Helmholtz machines and variational autoencoders, these RPM approximations introduce only minor bias, which may often vanish asymptotically. Furthermore, where the prior on latents is intractable the RPM may be combined effectively with standard probabilistic techniques such as variational Bayes. We demonstrate the model in high dimensional data settings, including a form of weakly supervised learning on MNIST digits and the discovery of latent maps from sensory observations. The RPM provides an effective way to discover, represent and reason probabilistically about the latent structure underlying observational data, functions which are critical to both animal and artificial intelligence.
Amortised Inference in Structured Generative Models with Explaining Away
Sep 12, 2022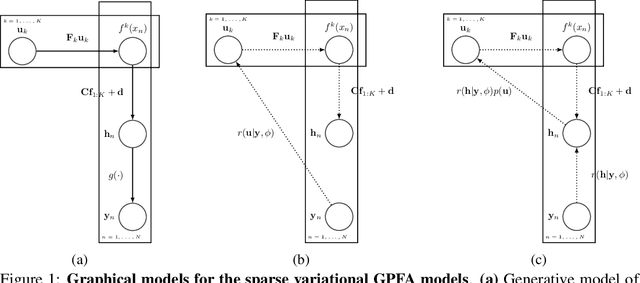
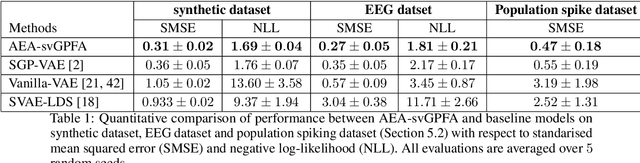
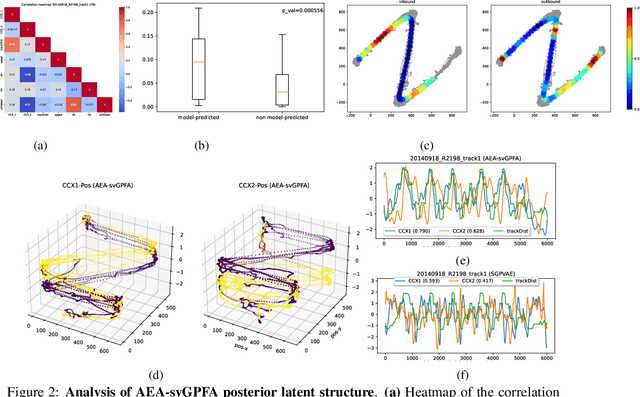
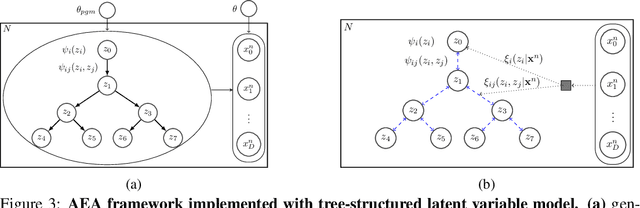
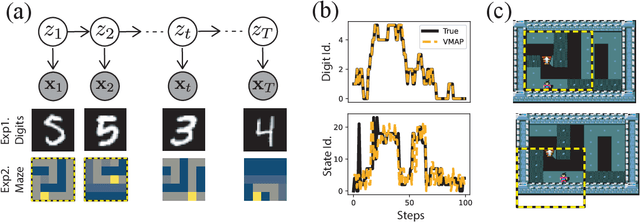
Abstract:A key goal of unsupervised learning is to go beyond density estimation and sample generation to reveal the structure inherent within observed data. Such structure can be expressed in the pattern of interactions between explanatory latent variables captured through a probabilistic graphical model. Although the learning of structured graphical models has a long history, much recent work in unsupervised modelling has instead emphasised flexible deep-network-based generation, either transforming independent latent generators to model complex data or assuming that distinct observed variables are derived from different latent nodes. Here, we extend the output of amortised variational inference to incorporate structured factors over multiple variables, able to capture the observation-induced posterior dependence between latents that results from "explaining away" and thus allow complex observations to depend on multiple nodes of a structured graph. We show that appropriately parameterised factors can be combined efficiently with variational message passing in elaborate graphical structures. We instantiate the framework based on Gaussian Process Factor Analysis models, and empirically evaluate its improvement over existing methods on synthetic data with known generative processes. We then fit the structured model to high-dimensional neural spiking time-series from the hippocampus of freely moving rodents, demonstrating that the model identifies latent signals that correlate with behavioural covariates.
Multitaper Spectral Estimation HDP-HMMs for EEG Sleep Inference
May 18, 2018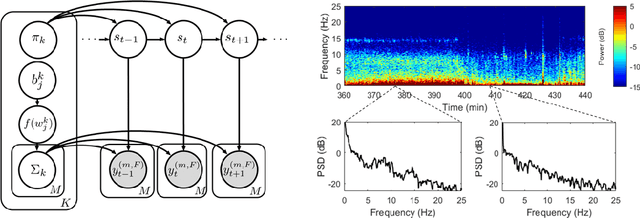
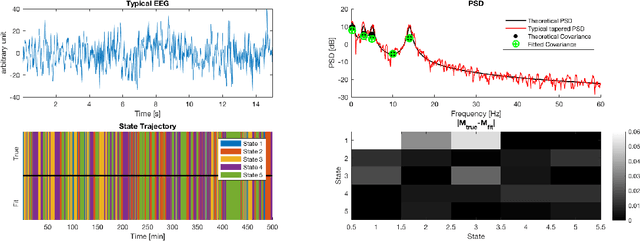
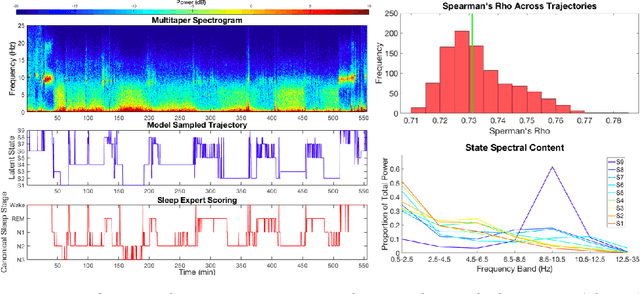
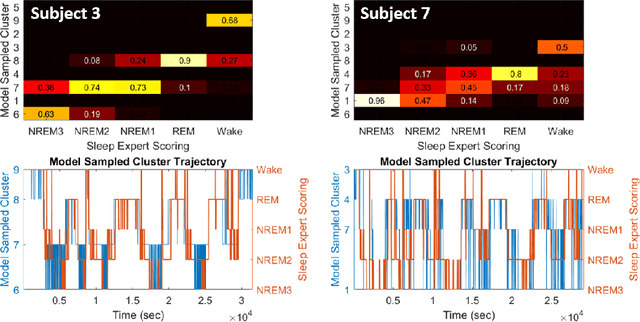
Abstract:Electroencephalographic (EEG) monitoring of neural activity is widely used for sleep disorder diagnostics and research. The standard of care is to manually classify 30-second epochs of EEG time-domain traces into 5 discrete sleep stages. Unfortunately, this scoring process is subjective and time-consuming, and the defined stages do not capture the heterogeneous landscape of healthy and clinical neural dynamics. This motivates the search for a data-driven and principled way to identify the number and composition of salient, reoccurring brain states present during sleep. To this end, we propose a Hierarchical Dirichlet Process Hidden Markov Model (HDP-HMM), combined with wide-sense stationary (WSS) time series spectral estimation to construct a generative model for personalized subject sleep states. In addition, we employ multitaper spectral estimation to further reduce the large variance of the spectral estimates inherent to finite-length EEG measurements. By applying our method to both simulated and human sleep data, we arrive at three main results: 1) a Bayesian nonparametric automated algorithm that recovers general temporal dynamics of sleep, 2) identification of subject-specific "microstates" within canonical sleep stages, and 3) discovery of stage-dependent sub-oscillations with shared spectral signatures across subjects.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge