Andrew Song
Robot Metabolism: Towards machines that can grow by consuming other machines
Nov 17, 2024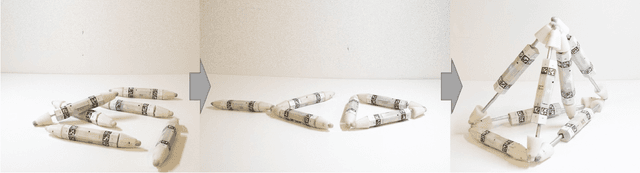
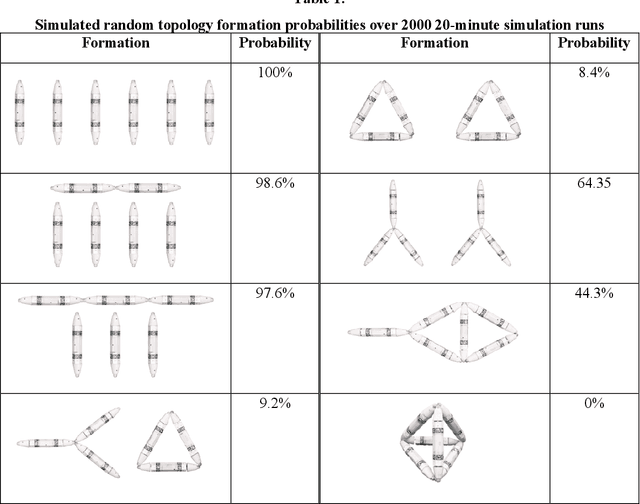
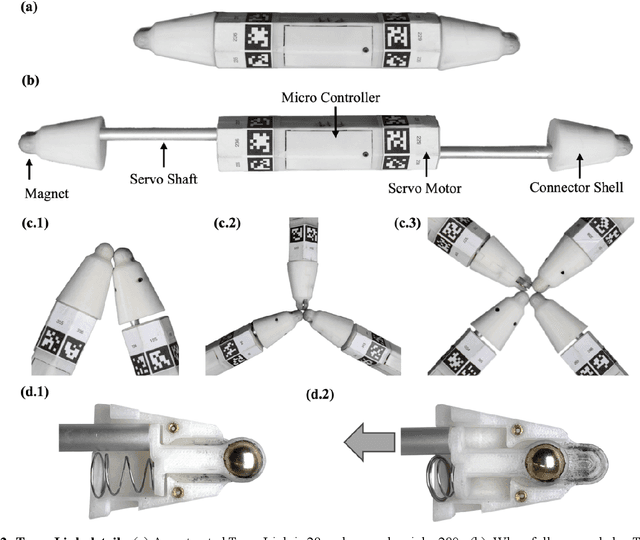
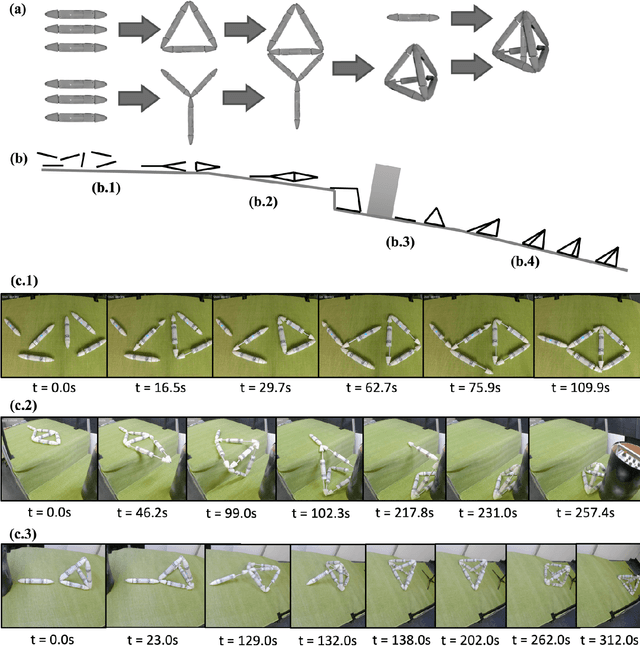
Abstract:Biological lifeforms can heal, grow, adapt, and reproduce -- abilities essential for sustained survival and development. In contrast, robots today are primarily monolithic machines with limited ability to self-repair, physically develop, or incorporate material from their environments. A key challenge to such physical adaptation has been that while robot minds are rapidly evolving new behaviors through AI, their bodies remain closed systems, unable to systematically integrate new material to grow or heal. We argue that open-ended physical adaptation is only possible when robots are designed using only a small repertoire of simple modules. This allows machines to mechanically adapt by consuming parts from other machines or their surroundings and shedding broken components. We demonstrate this principle using a truss modular robot platform composed of one-dimensional actuated bars. We show how robots in this space can grow bigger, faster, and more capable by consuming materials from their environment and from other robots. We suggest that machine metabolic processes akin to the one demonstrated here will be an essential part of any sustained future robot ecology.
Multitaper Spectral Estimation HDP-HMMs for EEG Sleep Inference
May 18, 2018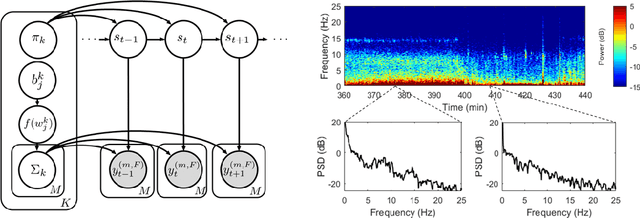
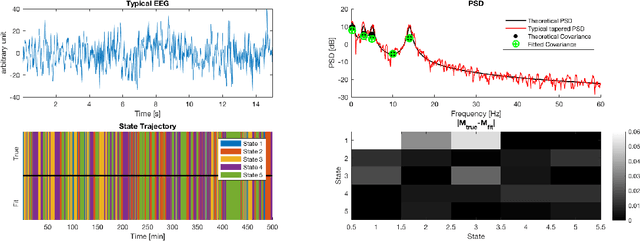
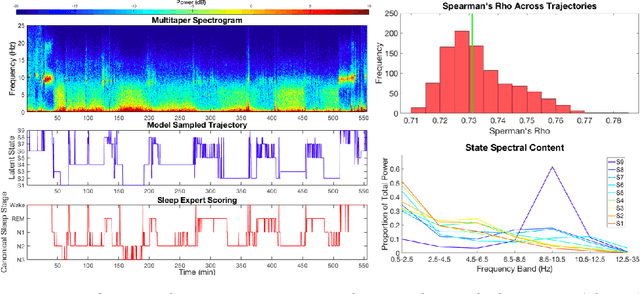
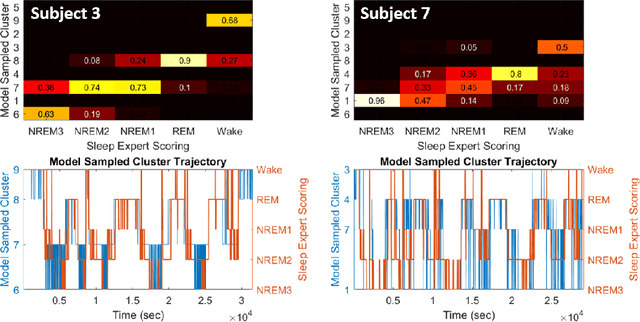
Abstract:Electroencephalographic (EEG) monitoring of neural activity is widely used for sleep disorder diagnostics and research. The standard of care is to manually classify 30-second epochs of EEG time-domain traces into 5 discrete sleep stages. Unfortunately, this scoring process is subjective and time-consuming, and the defined stages do not capture the heterogeneous landscape of healthy and clinical neural dynamics. This motivates the search for a data-driven and principled way to identify the number and composition of salient, reoccurring brain states present during sleep. To this end, we propose a Hierarchical Dirichlet Process Hidden Markov Model (HDP-HMM), combined with wide-sense stationary (WSS) time series spectral estimation to construct a generative model for personalized subject sleep states. In addition, we employ multitaper spectral estimation to further reduce the large variance of the spectral estimates inherent to finite-length EEG measurements. By applying our method to both simulated and human sleep data, we arrive at three main results: 1) a Bayesian nonparametric automated algorithm that recovers general temporal dynamics of sleep, 2) identification of subject-specific "microstates" within canonical sleep stages, and 3) discovery of stage-dependent sub-oscillations with shared spectral signatures across subjects.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge