Fuad Noman
SFC-GAN: A Generative Adversarial Network for Brain Functional and Structural Connectome Translation
Jan 13, 2025Abstract:Modern brain imaging technologies have enabled the detailed reconstruction of human brain connectomes, capturing structural connectivity (SC) from diffusion MRI and functional connectivity (FC) from functional MRI. Understanding the intricate relationships between SC and FC is vital for gaining deeper insights into the brain's functional and organizational mechanisms. However, obtaining both SC and FC modalities simultaneously remains challenging, hindering comprehensive analyses. Existing deep generative models typically focus on synthesizing a single modality or unidirectional translation between FC and SC, thereby missing the potential benefits of bi-directional translation, especially in scenarios where only one connectome is available. Therefore, we propose Structural-Functional Connectivity GAN (SFC-GAN), a novel framework for bidirectional translation between SC and FC. This approach leverages the CycleGAN architecture, incorporating convolutional layers to effectively capture the spatial structures of brain connectomes. To preserve the topological integrity of these connectomes, we employ a structure-preserving loss that guides the model in capturing both global and local connectome patterns while maintaining symmetry. Our framework demonstrates superior performance in translating between SC and FC, outperforming baseline models in similarity and graph property evaluations compared to ground truth data, each translated modality can be effectively utilized for downstream classification.
Dynamic MRI reconstruction using low-rank plus sparse decomposition with smoothness regularization
Jan 30, 2024



Abstract:The low-rank plus sparse (L+S) decomposition model has enabled better reconstruction of dynamic magnetic resonance imaging (dMRI) with separation into background (L) and dynamic (S) component. However, use of low-rank prior alone may not fully explain the slow variations or smoothness of the background part at the local scale. In this paper, we propose a smoothness-regularized L+S (SR-L+S) model for dMRI reconstruction from highly undersampled k-t-space data. We exploit joint low-rank and smooth priors on the background component of dMRI to better capture both its global and local temporal correlated structures. Extending the L+S formulation, the low-rank property is encoded by the nuclear norm, while the smoothness by a general \ell_{p}-norm penalty on the local differences of the columns of L. The additional smoothness regularizer can promote piecewise local consistency between neighboring frames. By smoothing out the noise and dynamic activities, it allows accurate recovery of the background part, and subsequently more robust dMRI reconstruction. Extensive experiments on multi-coil cardiac and synthetic data shows that the SR-L+S model outp
A Deep Probabilistic Spatiotemporal Framework for Dynamic Graph Representation Learning with Application to Brain Disorder Identification
Feb 16, 2023Abstract:Recent applications of pattern recognition techniques on brain connectome classification using functional connectivity (FC) neglect the non-Euclidean topology and causal dynamics of brain connectivity across time. In this paper, a deep probabilistic spatiotemporal framework developed based on variational Bayes (DSVB) is proposed to learn time-varying topological structures in dynamic brain FC networks for autism spectrum disorder (ASD) identification. The proposed framework incorporates a spatial-aware recurrent neural network to capture rich spatiotemporal patterns across dynamic FC networks, followed by a fully-connected neural network to exploit these learned patterns for subject-level classification. To overcome model overfitting on limited training datasets, an adversarial training strategy is introduced to learn graph embedding models that generalize well to unseen brain networks. Evaluation on the ABIDE resting-state functional magnetic resonance imaging dataset shows that our proposed framework significantly outperformed state-of-the-art methods in identifying ASD. Dynamic FC analyses with DSVB learned embeddings reveal apparent group difference between ASD and healthy controls in network profiles and switching dynamics of brain states.
Graph-Regularized Manifold-Aware Conditional Wasserstein GAN for Brain Functional Connectivity Generation
Dec 10, 2022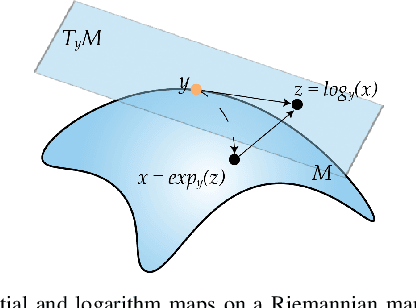
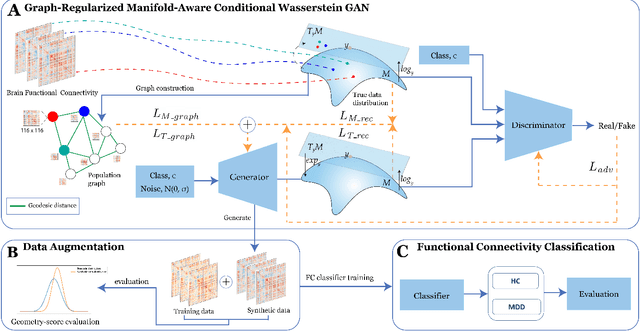
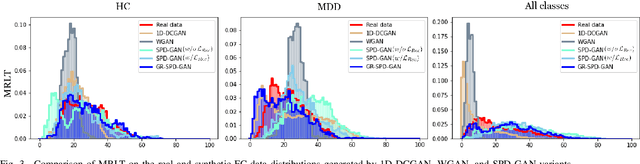
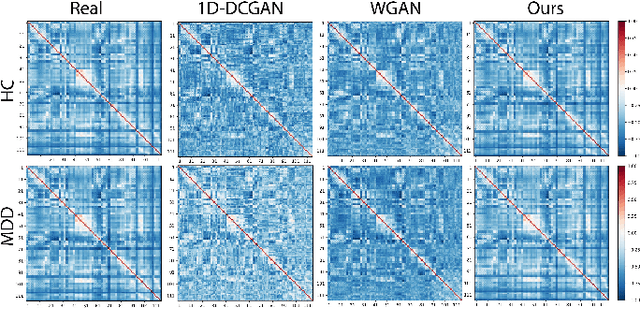
Abstract:Common measures of brain functional connectivity (FC) including covariance and correlation matrices are semi-positive definite (SPD) matrices residing on a cone-shape Riemannian manifold. Despite its remarkable success for Euclidean-valued data generation, use of standard generative adversarial networks (GANs) to generate manifold-valued FC data neglects its inherent SPD structure and hence the inter-relatedness of edges in real FC. We propose a novel graph-regularized manifold-aware conditional Wasserstein GAN (GR-SPD-GAN) for FC data generation on the SPD manifold that can preserve the global FC structure. Specifically, we optimize a generalized Wasserstein distance between the real and generated SPD data under an adversarial training, conditioned on the class labels. The resulting generator can synthesize new SPD-valued FC matrices associated with different classes of brain networks, e.g., brain disorder or healthy control. Furthermore, we introduce additional population graph-based regularization terms on both the SPD manifold and its tangent space to encourage the generator to respect the inter-subject similarity of FC patterns in the real data. This also helps in avoiding mode collapse and produces more stable GAN training. Evaluated on resting-state functional magnetic resonance imaging (fMRI) data of major depressive disorder (MDD), qualitative and quantitative results show that the proposed GR-SPD-GAN clearly outperforms several state-of-the-art GANs in generating more realistic fMRI-based FC samples. When applied to FC data augmentation for MDD identification, classification models trained on augmented data generated by our approach achieved the largest margin of improvement in classification accuracy among the competing GANs over baselines without data augmentation.
Graph Autoencoders for Embedding Learning in Brain Networks and Major Depressive Disorder Identification
Jul 27, 2021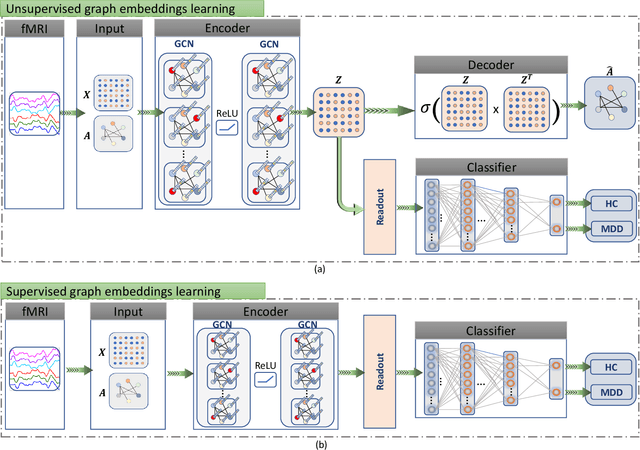
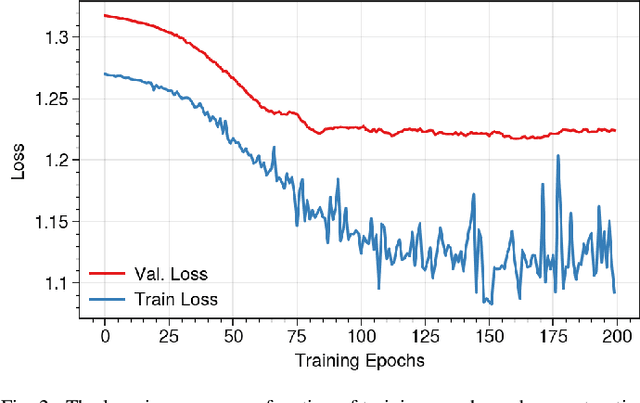
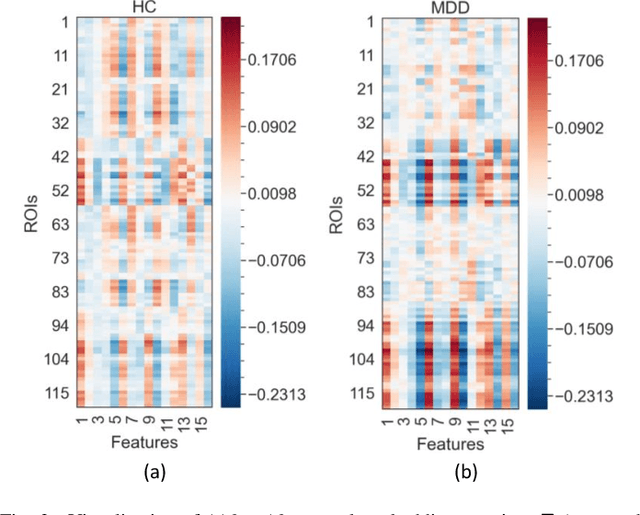
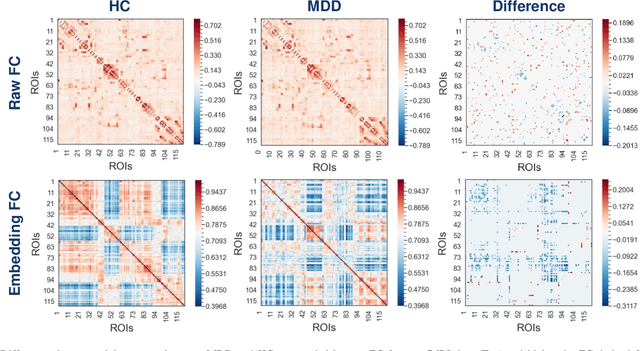
Abstract:Brain functional connectivity (FC) reveals biomarkers for identification of various neuropsychiatric disorders. Recent application of deep neural networks (DNNs) to connectome-based classification mostly relies on traditional convolutional neural networks using input connectivity matrices on a regular Euclidean grid. We propose a graph deep learning framework to incorporate the non-Euclidean information about graph structure for classifying functional magnetic resonance imaging (fMRI)- derived brain networks in major depressive disorder (MDD). We design a novel graph autoencoder (GAE) architecture based on the graph convolutional networks (GCNs) to embed the topological structure and node content of large-sized fMRI networks into low-dimensional latent representations. In network construction, we employ the Ledoit-Wolf (LDW) shrinkage method to estimate the high-dimensional FC metrics efficiently from fMRI data. We consider both supervised and unsupervised approaches for the graph embedded learning. The learned embeddings are then used as feature inputs for a deep fully-connected neural network (FCNN) to discriminate MDD from healthy controls. Evaluated on a resting-state fMRI MDD dataset with 43 subjects, results show that the proposed GAE-FCNN model significantly outperforms several state-of-the-art DNN methods for brain connectome classification, achieving accuracy of 72.50% using the LDW-FC metrics as node features. The graph embeddings of fMRI FC networks learned by the GAE also reveal apparent group differences between MDD and HC. Our new framework demonstrates feasibility of learning graph embeddings on brain networks to provide discriminative information for diagnosis of brain disorders.
Classification of EEG-Based Brain Connectivity Networks in Schizophrenia Using a Multi-Domain Connectome Convolutional Neural Network
Mar 21, 2019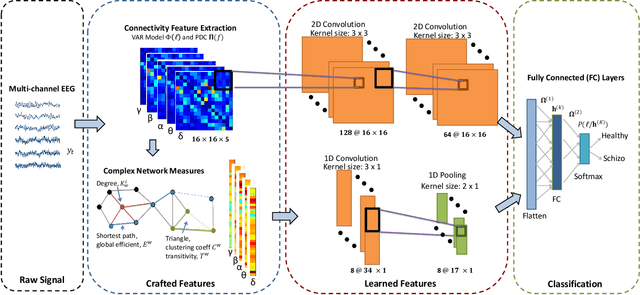
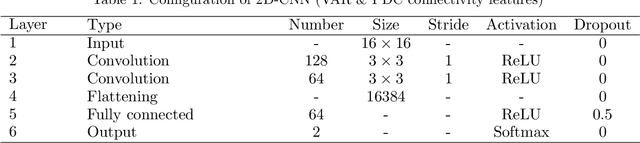
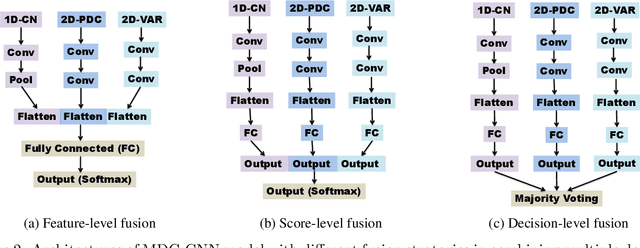

Abstract:We exploit altered patterns in brain functional connectivity as features for automatic discriminative analysis of neuropsychiatric patients. Deep learning methods have been introduced to functional network classification only very recently for fMRI, and the proposed architectures essentially focused on a single type of connectivity measure. We propose a deep convolutional neural network (CNN) framework for classification of electroencephalogram (EEG)-derived brain connectome in schizophrenia (SZ). To capture complementary aspects of disrupted connectivity in SZ, we explore combination of various connectivity features consisting of time and frequency-domain metrics of effective connectivity based on vector autoregressive model and partial directed coherence, and complex network measures of network topology. We design a novel multi-domain connectome CNN (MDC-CNN) based on a parallel ensemble of 1D and 2D CNNs to integrate the features from various domains and dimensions using different fusion strategies. Hierarchical latent representations learned by the multiple convolutional layers from EEG connectivity reveal apparent group differences between SZ and healthy controls (HC). Results on a large resting-state EEG dataset show that the proposed CNNs significantly outperform traditional support vector machine classifiers. The MDC-CNN with combined connectivity features further improves performance over single-domain CNNs using individual features, achieving remarkable accuracy of $93.06\%$ with a decision-level fusion. The proposed MDC-CNN by integrating information from diverse brain connectivity descriptors is able to accurately discriminate SZ from HC. The new framework is potentially useful for developing diagnostic tools for SZ and other disorders.
Short-segment heart sound classification using an ensemble of deep convolutional neural networks
Oct 27, 2018



Abstract:This paper proposes a framework based on deep convolutional neural networks (CNNs) for automatic heart sound classification using short-segments of individual heart beats. We design a 1D-CNN that directly learns features from raw heart-sound signals, and a 2D-CNN that takes inputs of two- dimensional time-frequency feature maps based on Mel-frequency cepstral coefficients (MFCC). We further develop a time-frequency CNN ensemble (TF-ECNN) combining the 1D-CNN and 2D-CNN based on score-level fusion of the class probabilities. On the large PhysioNet CinC challenge 2016 database, the proposed CNN models outperformed traditional classifiers based on support vector machine and hidden Markov models with various hand-crafted time- and frequency-domain features. Best classification scores with 89.22% accuracy and 89.94% sensitivity were achieved by the ECNN, and 91.55% specificity and 88.82% modified accuracy by the 2D-CNN alone on the test set.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge