Raphaël C. -W. Phan
Causal-Ex: Causal Graph-based Micro and Macro Expression Spotting
Mar 12, 2025Abstract:Detecting concealed emotions within apparently normal expressions is crucial for identifying potential mental health issues and facilitating timely support and intervention. The task of spotting macro and micro-expressions involves predicting the emotional timeline within a video, accomplished by identifying the onset, apex, and offset frames of the displayed emotions. Utilizing foundational facial muscle movement cues, known as facial action units, boosts the accuracy. However, an overlooked challenge from previous research lies in the inadvertent integration of biases into the training model. These biases arising from datasets can spuriously link certain action unit movements to particular emotion classes. We tackle this issue by novel replacement of action unit adjacency information with the action unit causal graphs. This approach aims to identify and eliminate undesired spurious connections, retaining only unbiased information for classification. Our model, named Causal-Ex (Causal-based Expression spotting), employs a rapid causal inference algorithm to construct a causal graph of facial action units. This enables us to select causally relevant facial action units. Our work demonstrates improvement in overall F1-scores compared to state-of-the-art approaches with 0.388 on CAS(ME)^2 and 0.3701 on SAMM-Long Video datasets.
PK-YOLO: Pretrained Knowledge Guided YOLO for Brain Tumor Detection in Multiplanar MRI Slices
Oct 29, 2024Abstract:Brain tumor detection in multiplane Magnetic Resonance Imaging (MRI) slices is a challenging task due to the various appearances and relationships in the structure of the multiplane images. In this paper, we propose a new You Only Look Once (YOLO)-based detection model that incorporates Pretrained Knowledge (PK), called PK-YOLO, to improve the performance for brain tumor detection in multiplane MRI slices. To our best knowledge, PK-YOLO is the first pretrained knowledge guided YOLO-based object detector. The main components of the new method are a pretrained pure lightweight convolutional neural network-based backbone via sparse masked modeling, a YOLO architecture with the pretrained backbone, and a regression loss function for improving small object detection. The pretrained backbone allows for feature transferability of object queries on individual plane MRI slices into the model encoders, and the learned domain knowledge base can improve in-domain detection. The improved loss function can further boost detection performance on small-size brain tumors in multiplanar two-dimensional MRI slices. Experimental results show that the proposed PK-YOLO achieves competitive performance on the multiplanar MRI brain tumor detection datasets compared to state-of-the-art YOLO-like and DETR-like object detectors. The code is available at https://github.com/mkang315/PK-YOLO.
A Multimodal Feature Distillation with CNN-Transformer Network for Brain Tumor Segmentation with Incomplete Modalities
Apr 22, 2024



Abstract:Existing brain tumor segmentation methods usually utilize multiple Magnetic Resonance Imaging (MRI) modalities in brain tumor images for segmentation, which can achieve better segmentation performance. However, in clinical applications, some modalities are missing due to resource constraints, leading to severe degradation in the performance of methods applying complete modality segmentation. In this paper, we propose a Multimodal feature distillation with Convolutional Neural Network (CNN)-Transformer hybrid network (MCTSeg) for accurate brain tumor segmentation with missing modalities. We first design a Multimodal Feature Distillation (MFD) module to distill feature-level multimodal knowledge into different unimodality to extract complete modality information. We further develop a Unimodal Feature Enhancement (UFE) module to model the relationship between global and local information semantically. Finally, we build a Cross-Modal Fusion (CMF) module to explicitly align the global correlations among different modalities even when some modalities are missing. Complementary features within and across different modalities are refined via the CNN-Transformer hybrid architectures in both the UFE and CMF modules, where local and global dependencies are both captured. Our ablation study demonstrates the importance of the proposed modules with CNN-Transformer networks and the convolutional blocks in Transformer for improving the performance of brain tumor segmentation with missing modalities. Extensive experiments on the BraTS2018 and BraTS2020 datasets show that the proposed MCTSeg framework outperforms the state-of-the-art methods in missing modalities cases. Our code is available at: https://github.com/mkang315/MCTSeg.
Dynamic MRI reconstruction using low-rank plus sparse decomposition with smoothness regularization
Jan 30, 2024



Abstract:The low-rank plus sparse (L+S) decomposition model has enabled better reconstruction of dynamic magnetic resonance imaging (dMRI) with separation into background (L) and dynamic (S) component. However, use of low-rank prior alone may not fully explain the slow variations or smoothness of the background part at the local scale. In this paper, we propose a smoothness-regularized L+S (SR-L+S) model for dMRI reconstruction from highly undersampled k-t-space data. We exploit joint low-rank and smooth priors on the background component of dMRI to better capture both its global and local temporal correlated structures. Extending the L+S formulation, the low-rank property is encoded by the nuclear norm, while the smoothness by a general \ell_{p}-norm penalty on the local differences of the columns of L. The additional smoothness regularizer can promote piecewise local consistency between neighboring frames. By smoothing out the noise and dynamic activities, it allows accurate recovery of the background part, and subsequently more robust dMRI reconstruction. Extensive experiments on multi-coil cardiac and synthetic data shows that the SR-L+S model outp
ASF-YOLO: A Novel YOLO Model with Attentional Scale Sequence Fusion for Cell Instance Segmentation
Dec 11, 2023Abstract:We propose a novel Attentional Scale Sequence Fusion based You Only Look Once (YOLO) framework (ASF-YOLO) which combines spatial and scale features for accurate and fast cell instance segmentation. Built on the YOLO segmentation framework, we employ the Scale Sequence Feature Fusion (SSFF) module to enhance the multi-scale information extraction capability of the network, and the Triple Feature Encoder (TPE) module to fuse feature maps of different scales to increase detailed information. We further introduce a Channel and Position Attention Mechanism (CPAM) to integrate both the SSFF and TPE modules, which focus on informative channels and spatial position-related small objects for improved detection and segmentation performance. Experimental validations on two cell datasets show remarkable segmentation accuracy and speed of the proposed ASF-YOLO model. It achieves a box mAP of 0.91, mask mAP of 0.887, and an inference speed of 47.3 FPS on the 2018 Data Science Bowl dataset, outperforming the state-of-the-art methods. The source code is available at https://github.com/mkang315/ASF-YOLO.
BGF-YOLO: Enhanced YOLOv8 with Multiscale Attentional Feature Fusion for Brain Tumor Detection
Sep 25, 2023Abstract:You Only Look Once (YOLO)-based object detectors have shown remarkable accuracy for automated brain tumor detection. In this paper, we develop a novel BGF-YOLO architecture by incorporating Bi-level Routing Attention (BRA), Generalized feature pyramid networks (GFPN), and Fourth detecting head into YOLOv8. BGF-YOLO contains an attention mechanism to focus more on important features, and feature pyramid networks to enrich feature representation by merging high-level semantic features with spatial details. Furthermore, we investigate the effect of different attention mechanisms and feature fusions, detection head architectures on brain tumor detection accuracy. Experimental results show that BGF-YOLO gives a 4.7% absolute increase of mAP$_{50}$ compared to YOLOv8x, and achieves state-of-the-art on the brain tumor detection dataset Br35H. The code is available at https://github.com/mkang315/BGF-YOLO.
RCS-YOLO: A Fast and High-Accuracy Object Detector for Brain Tumor Detection
Jul 31, 2023Abstract:With an excellent balance between speed and accuracy, cutting-edge YOLO frameworks have become one of the most efficient algorithms for object detection. However, the performance of using YOLO networks is scarcely investigated in brain tumor detection. We propose a novel YOLO architecture with Reparameterized Convolution based on channel Shuffle (RCS-YOLO). We present RCS and a One-Shot Aggregation of RCS (RCS-OSA), which link feature cascade and computation efficiency to extract richer information and reduce time consumption. Experimental results on the brain tumor dataset Br35H show that the proposed model surpasses YOLOv6, YOLOv7, and YOLOv8 in speed and accuracy. Notably, compared with YOLOv7, the precision of RCS-YOLO improves by 2.6%, and the inference speed by 60% at 114.8 images detected per second (FPS). Our proposed RCS-YOLO achieves state-of-the-art performance on the brain tumor detection task. The code is available at https://github.com/mkang315/RCS-YOLO.
Graph-Regularized Manifold-Aware Conditional Wasserstein GAN for Brain Functional Connectivity Generation
Dec 10, 2022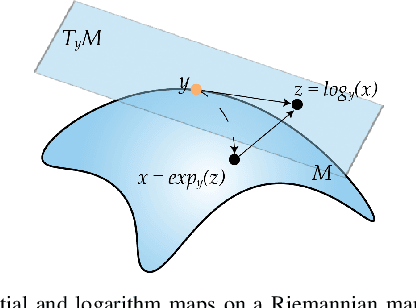
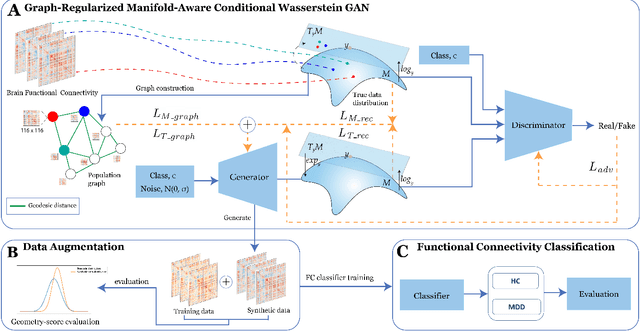
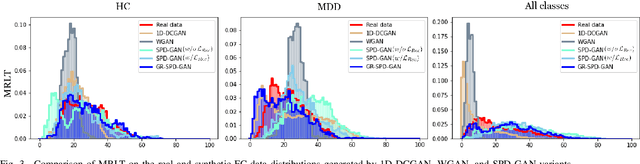
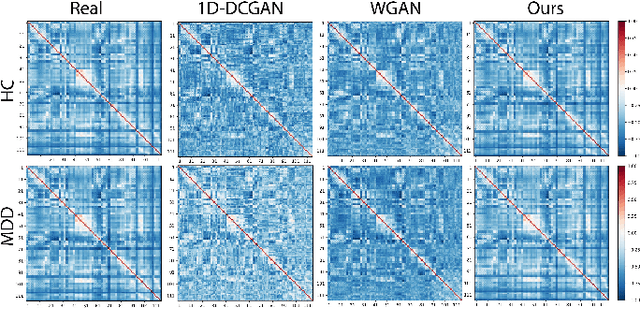
Abstract:Common measures of brain functional connectivity (FC) including covariance and correlation matrices are semi-positive definite (SPD) matrices residing on a cone-shape Riemannian manifold. Despite its remarkable success for Euclidean-valued data generation, use of standard generative adversarial networks (GANs) to generate manifold-valued FC data neglects its inherent SPD structure and hence the inter-relatedness of edges in real FC. We propose a novel graph-regularized manifold-aware conditional Wasserstein GAN (GR-SPD-GAN) for FC data generation on the SPD manifold that can preserve the global FC structure. Specifically, we optimize a generalized Wasserstein distance between the real and generated SPD data under an adversarial training, conditioned on the class labels. The resulting generator can synthesize new SPD-valued FC matrices associated with different classes of brain networks, e.g., brain disorder or healthy control. Furthermore, we introduce additional population graph-based regularization terms on both the SPD manifold and its tangent space to encourage the generator to respect the inter-subject similarity of FC patterns in the real data. This also helps in avoiding mode collapse and produces more stable GAN training. Evaluated on resting-state functional magnetic resonance imaging (fMRI) data of major depressive disorder (MDD), qualitative and quantitative results show that the proposed GR-SPD-GAN clearly outperforms several state-of-the-art GANs in generating more realistic fMRI-based FC samples. When applied to FC data augmentation for MDD identification, classification models trained on augmented data generated by our approach achieved the largest margin of improvement in classification accuracy among the competing GANs over baselines without data augmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge