Farhan Damani
Beyond the training set: an intuitive method for detecting distribution shift in model-based optimization
Nov 09, 2023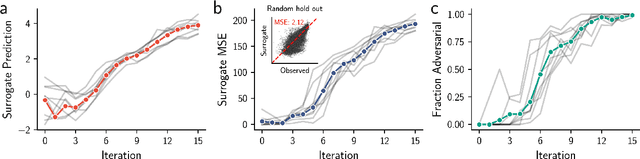
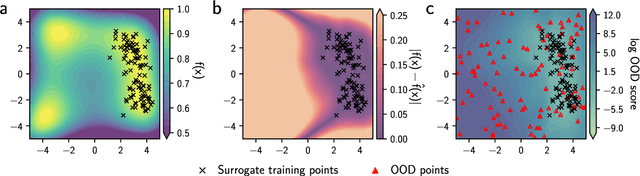
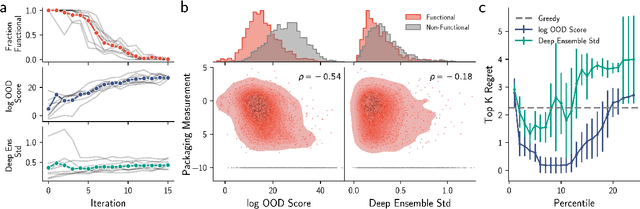
Abstract:Model-based optimization (MBO) is increasingly applied to design problems in science and engineering. A common scenario involves using a fixed training set to train models, with the goal of designing new samples that outperform those present in the training data. A major challenge in this setting is distribution shift, where the distributions of training and design samples are different. While some shift is expected, as the goal is to create better designs, this change can negatively affect model accuracy and subsequently, design quality. Despite the widespread nature of this problem, addressing it demands deep domain knowledge and artful application. To tackle this issue, we propose a straightforward method for design practitioners that detects distribution shifts. This method trains a binary classifier using knowledge of the unlabeled design distribution to separate the training data from the design data. The classifier's logit scores are then used as a proxy measure of distribution shift. We validate our method in a real-world application by running offline MBO and evaluate the effect of distribution shift on design quality. We find that the intensity of the shift in the design distribution varies based on the number of steps taken by the optimization algorithm, and our simple approach can identify these shifts. This enables users to constrain their search to regions where the model's predictions are reliable, thereby increasing the quality of designs.
Black Box Recursive Translations for Molecular Optimization
Dec 21, 2019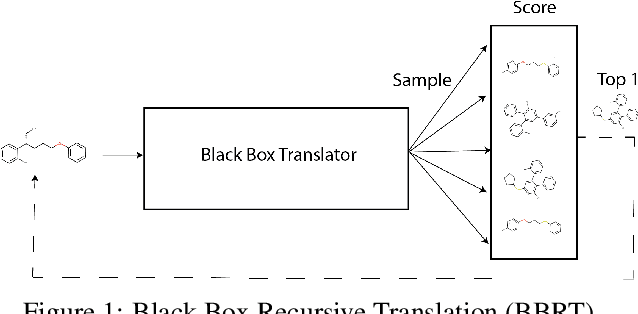
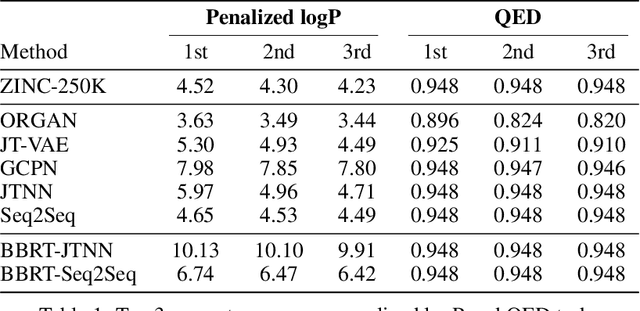
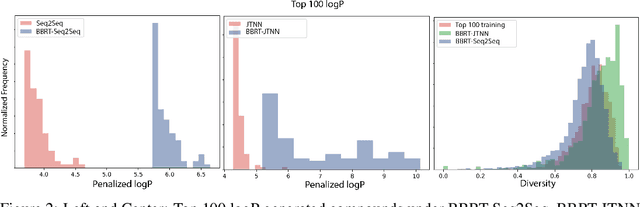
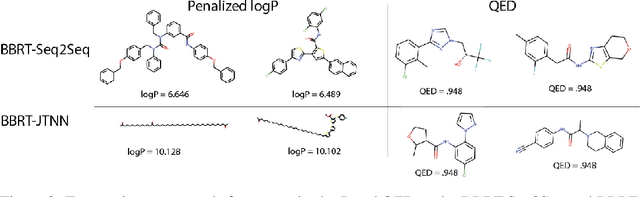
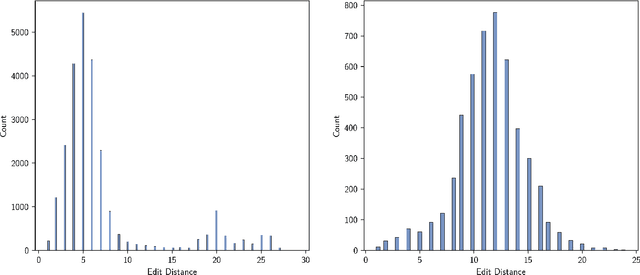
Abstract:Machine learning algorithms for generating molecular structures offer a promising new approach to drug discovery. We cast molecular optimization as a translation problem, where the goal is to map an input compound to a target compound with improved biochemical properties. Remarkably, we observe that when generated molecules are iteratively fed back into the translator, molecular compound attributes improve with each step. We show that this finding is invariant to the choice of translation model, making this a "black box" algorithm. We call this method Black Box Recursive Translation (BBRT), a new inference method for molecular property optimization. This simple, powerful technique operates strictly on the inputs and outputs of any translation model. We obtain new state-of-the-art results for molecular property optimization tasks using our simple drop-in replacement with well-known sequence and graph-based models. Our method provides a significant boost in performance relative to its non-recursive peers with just a simple "for" loop. Further, BBRT is highly interpretable, allowing users to map the evolution of newly discovered compounds from known starting points.
Discrete Object Generation with Reversible Inductive Construction
Jul 18, 2019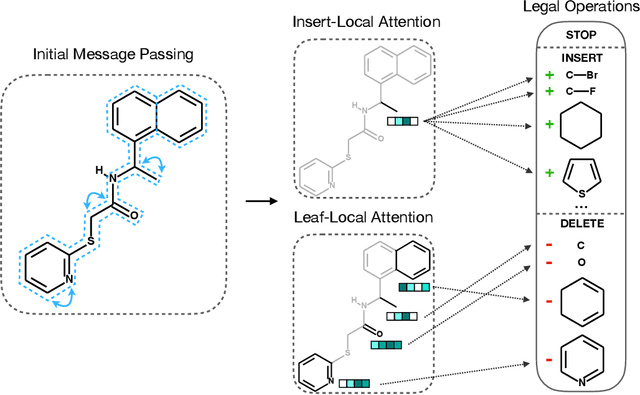
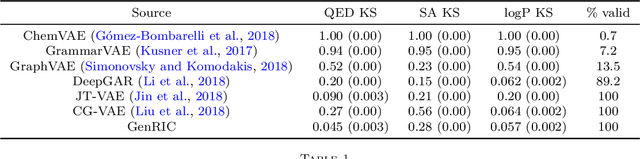
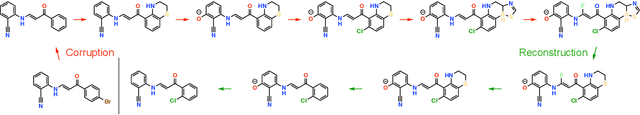
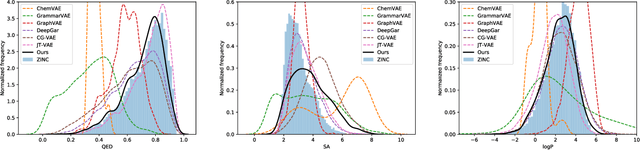
Abstract:The success of generative modeling in continuous domains has led to a surge of interest in generating discrete data such as molecules, source code, and graphs. However, construction histories for these discrete objects are typically not unique and so generative models must reason about intractably large spaces in order to learn. Additionally, structured discrete domains are often characterized by strict constraints on what constitutes a valid object and generative models must respect these requirements in order to produce useful novel samples. Here, we present a generative model for discrete objects employing a Markov chain where transitions are restricted to a set of local operations that preserve validity. Building off of generative interpretations of denoising autoencoders, the Markov chain alternates between producing 1) a sequence of corrupted objects that are valid but not from the data distribution, and 2) a learned reconstruction distribution that attempts to fix the corruptions while also preserving validity. This approach constrains the generative model to only produce valid objects, requires the learner to only discover local modifications to the objects, and avoids marginalization over an unknown and potentially large space of construction histories. We evaluate the proposed approach on two highly structured discrete domains, molecules and Laman graphs, and find that it compares favorably to alternative methods at capturing distributional statistics for a host of semantically relevant metrics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge