Stephen Malina
Michael Pokorny
Humanity's Last Exam
Jan 24, 2025Abstract:Benchmarks are important tools for tracking the rapid advancements in large language model (LLM) capabilities. However, benchmarks are not keeping pace in difficulty: LLMs now achieve over 90\% accuracy on popular benchmarks like MMLU, limiting informed measurement of state-of-the-art LLM capabilities. In response, we introduce Humanity's Last Exam (HLE), a multi-modal benchmark at the frontier of human knowledge, designed to be the final closed-ended academic benchmark of its kind with broad subject coverage. HLE consists of 3,000 questions across dozens of subjects, including mathematics, humanities, and the natural sciences. HLE is developed globally by subject-matter experts and consists of multiple-choice and short-answer questions suitable for automated grading. Each question has a known solution that is unambiguous and easily verifiable, but cannot be quickly answered via internet retrieval. State-of-the-art LLMs demonstrate low accuracy and calibration on HLE, highlighting a significant gap between current LLM capabilities and the expert human frontier on closed-ended academic questions. To inform research and policymaking upon a clear understanding of model capabilities, we publicly release HLE at https://lastexam.ai.
BioNeMo Framework: a modular, high-performance library for AI model development in drug discovery
Nov 15, 2024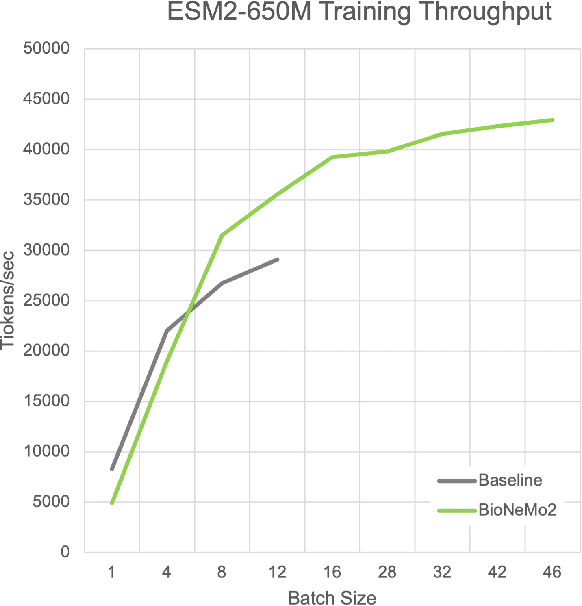
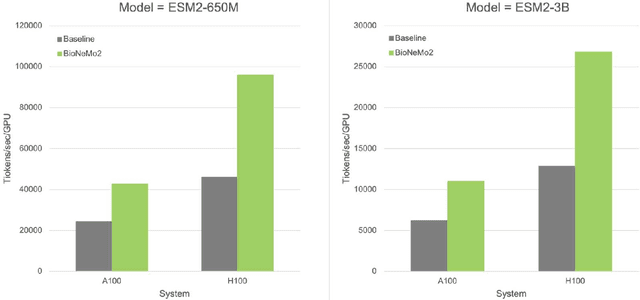
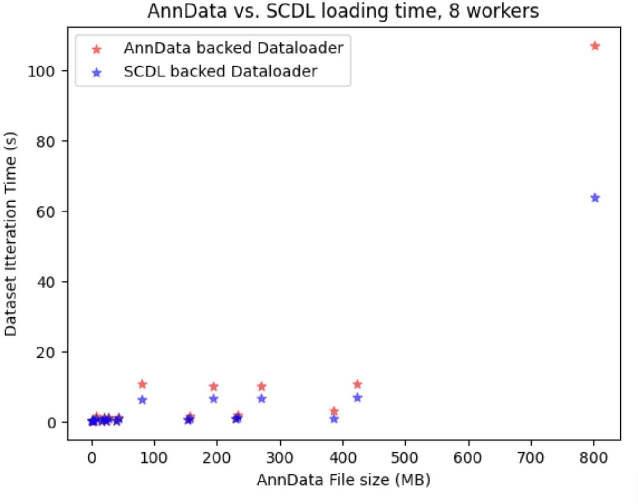
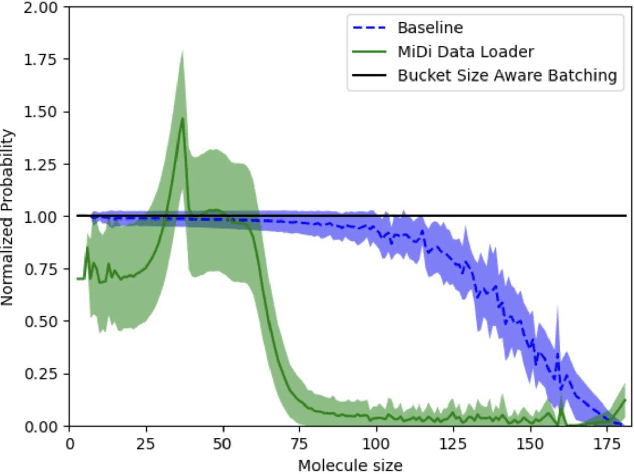
Abstract:Artificial Intelligence models encoding biology and chemistry are opening new routes to high-throughput and high-quality in-silico drug development. However, their training increasingly relies on computational scale, with recent protein language models (pLM) training on hundreds of graphical processing units (GPUs). We introduce the BioNeMo Framework to facilitate the training of computational biology and chemistry AI models across hundreds of GPUs. Its modular design allows the integration of individual components, such as data loaders, into existing workflows and is open to community contributions. We detail technical features of the BioNeMo Framework through use cases such as pLM pre-training and fine-tuning. On 256 NVIDIA A100s, BioNeMo Framework trains a three billion parameter BERT-based pLM on over one trillion tokens in 4.2 days. The BioNeMo Framework is open-source and free for everyone to use.
Beyond the training set: an intuitive method for detecting distribution shift in model-based optimization
Nov 09, 2023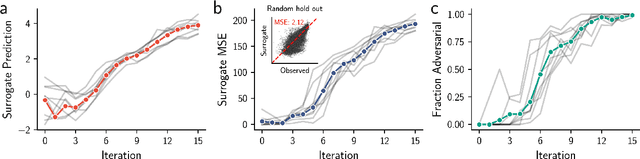
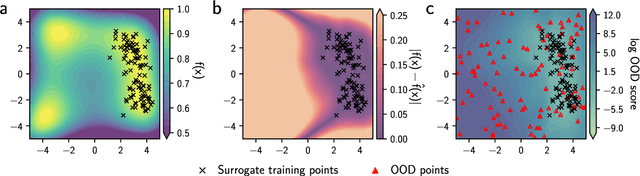
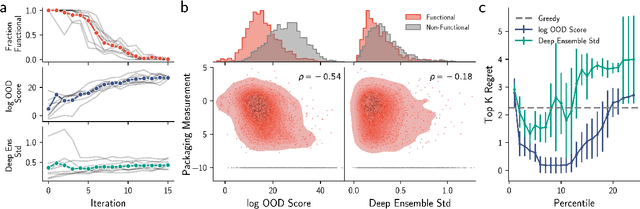
Abstract:Model-based optimization (MBO) is increasingly applied to design problems in science and engineering. A common scenario involves using a fixed training set to train models, with the goal of designing new samples that outperform those present in the training data. A major challenge in this setting is distribution shift, where the distributions of training and design samples are different. While some shift is expected, as the goal is to create better designs, this change can negatively affect model accuracy and subsequently, design quality. Despite the widespread nature of this problem, addressing it demands deep domain knowledge and artful application. To tackle this issue, we propose a straightforward method for design practitioners that detects distribution shifts. This method trains a binary classifier using knowledge of the unlabeled design distribution to separate the training data from the design data. The classifier's logit scores are then used as a proxy measure of distribution shift. We validate our method in a real-world application by running offline MBO and evaluate the effect of distribution shift on design quality. We find that the intensity of the shift in the design distribution varies based on the number of steps taken by the optimization algorithm, and our simple approach can identify these shifts. This enables users to constrain their search to regions where the model's predictions are reliable, thereby increasing the quality of designs.
Forecasting labels under distribution-shift for machine-guided sequence design
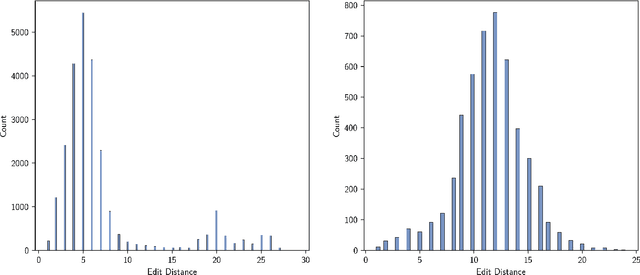
Nov 18, 2022Abstract:The ability to design and optimize biological sequences with specific functionalities would unlock enormous value in technology and healthcare. In recent years, machine learning-guided sequence design has progressed this goal significantly, though validating designed sequences in the lab or clinic takes many months and substantial labor. It is therefore valuable to assess the likelihood that a designed set contains sequences of the desired quality (which often lies outside the label distribution in our training data) before committing resources to an experiment. Forecasting, a prominent concept in many domains where feedback can be delayed (e.g. elections), has not been used or studied in the context of sequence design. Here we propose a method to guide decision-making that forecasts the performance of high-throughput libraries (e.g. containing $10^5$ unique variants) based on estimates provided by models, providing a posterior for the distribution of labels in the library. We show that our method outperforms baselines that naively use model scores to estimate library performance, which are the only tool available today for this purpose.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge