Eu Wern Teh
PrismAvatar: Real-time animated 3D neural head avatars on edge devices
Feb 10, 2025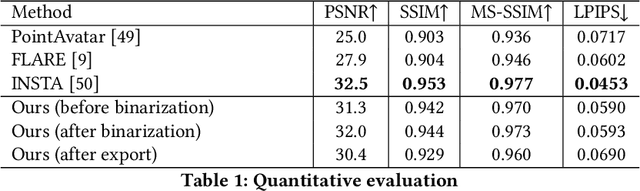


Abstract:We present PrismAvatar: a 3D head avatar model which is designed specifically to enable real-time animation and rendering on resource-constrained edge devices, while still enjoying the benefits of neural volumetric rendering at training time. By integrating a rigged prism lattice with a 3D morphable head model, we use a hybrid rendering model to simultaneously reconstruct a mesh-based head and a deformable NeRF model for regions not represented by the 3DMM. We then distill the deformable NeRF into a rigged mesh and neural textures, which can be animated and rendered efficiently within the constraints of the traditional triangle rendering pipeline. In addition to running at 60 fps with low memory usage on mobile devices, we find that our trained models have comparable quality to state-of-the-art 3D avatar models on desktop devices.
3D Face Tracking from 2D Video through Iterative Dense UV to Image Flow
Apr 15, 2024



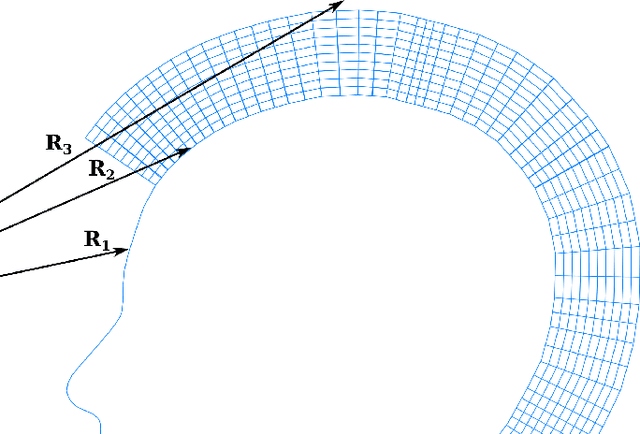
Abstract:When working with 3D facial data, improving fidelity and avoiding the uncanny valley effect is critically dependent on accurate 3D facial performance capture. Because such methods are expensive and due to the widespread availability of 2D videos, recent methods have focused on how to perform monocular 3D face tracking. However, these methods often fall short in capturing precise facial movements due to limitations in their network architecture, training, and evaluation processes. Addressing these challenges, we propose a novel face tracker, FlowFace, that introduces an innovative 2D alignment network for dense per-vertex alignment. Unlike prior work, FlowFace is trained on high-quality 3D scan annotations rather than weak supervision or synthetic data. Our 3D model fitting module jointly fits a 3D face model from one or many observations, integrating existing neutral shape priors for enhanced identity and expression disentanglement and per-vertex deformations for detailed facial feature reconstruction. Additionally, we propose a novel metric and benchmark for assessing tracking accuracy. Our method exhibits superior performance on both custom and publicly available benchmarks. We further validate the effectiveness of our tracker by generating high-quality 3D data from 2D videos, which leads to performance gains on downstream tasks.
Embracing Annotation Efficient Learning (AEL) for Digital Pathology and Natural Images
Dec 01, 2022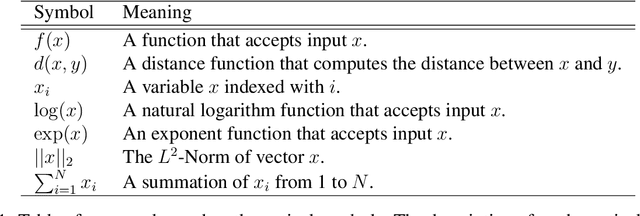
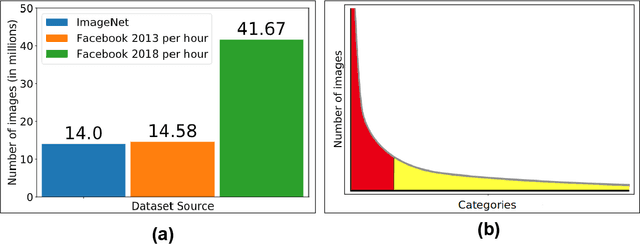

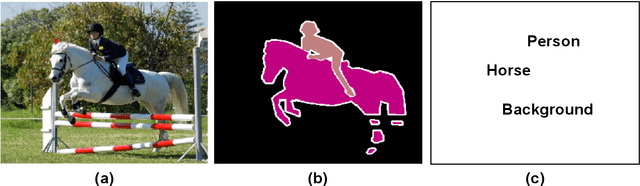
Abstract:Jitendra Malik once said, "Supervision is the opium of the AI researcher". Most deep learning techniques heavily rely on extreme amounts of human labels to work effectively. In today's world, the rate of data creation greatly surpasses the rate of data annotation. Full reliance on human annotations is just a temporary means to solve current closed problems in AI. In reality, only a tiny fraction of data is annotated. Annotation Efficient Learning (AEL) is a study of algorithms to train models effectively with fewer annotations. To thrive in AEL environments, we need deep learning techniques that rely less on manual annotations (e.g., image, bounding-box, and per-pixel labels), but learn useful information from unlabeled data. In this thesis, we explore five different techniques for handling AEL.
Understanding the impact of image and input resolution on deep digital pathology patch classifiers
Apr 29, 2022
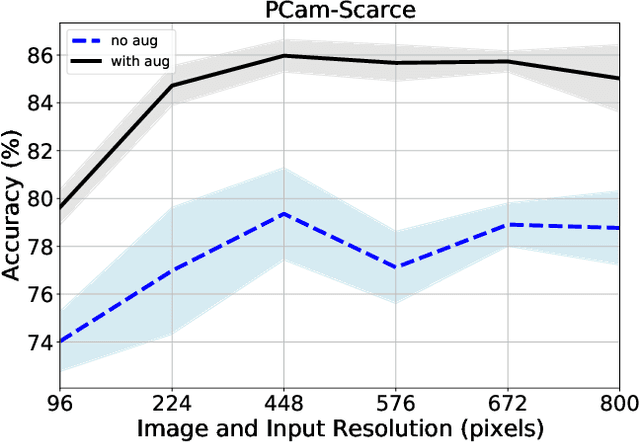
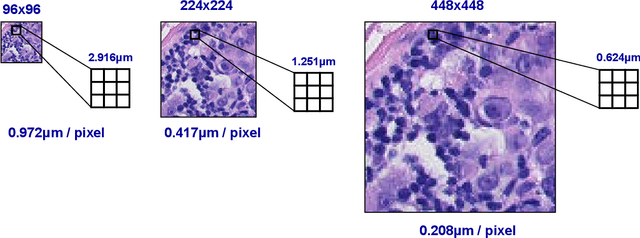
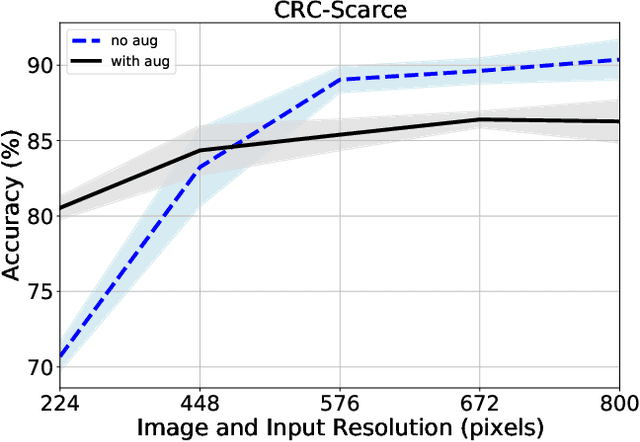
Abstract:We consider annotation efficient learning in Digital Pathology (DP), where expert annotations are expensive and thus scarce. We explore the impact of image and input resolution on DP patch classification performance. We use two cancer patch classification datasets PCam and CRC, to validate the results of our study. Our experiments show that patch classification performance can be improved by manipulating both the image and input resolution in annotation-scarce and annotation-rich environments. We show a positive correlation between the image and input resolution and the patch classification accuracy on both datasets. By exploiting the image and input resolution, our final model trained on < 1% of data performs equally well compared to the model trained on 100% of data in the original image resolution on the PCam dataset.
Learning with Less Labels in Digital Pathology via Scribble Supervision from Natural Images
Jan 20, 2022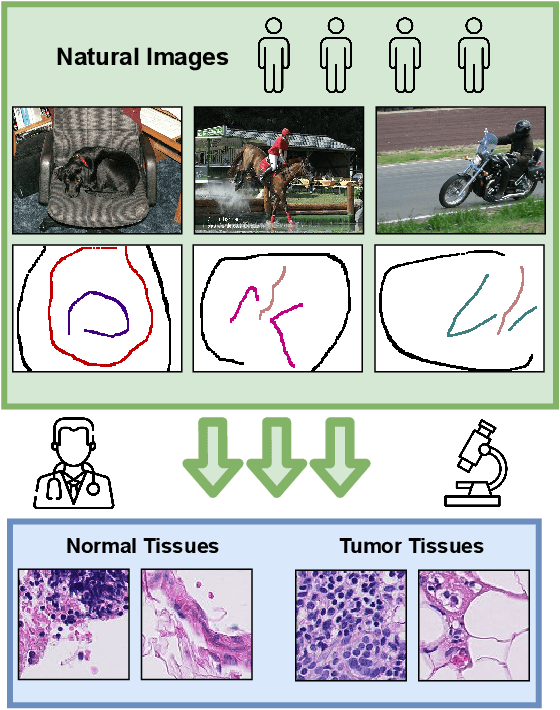
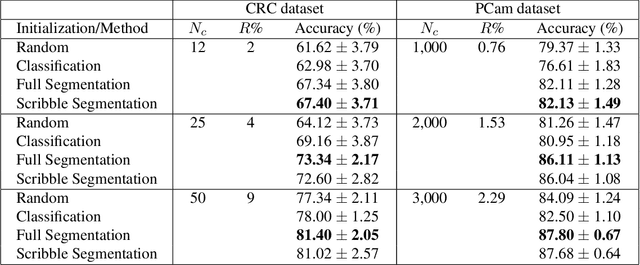
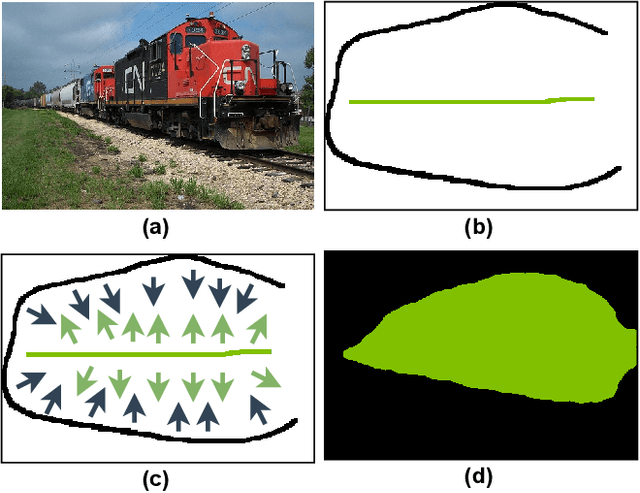
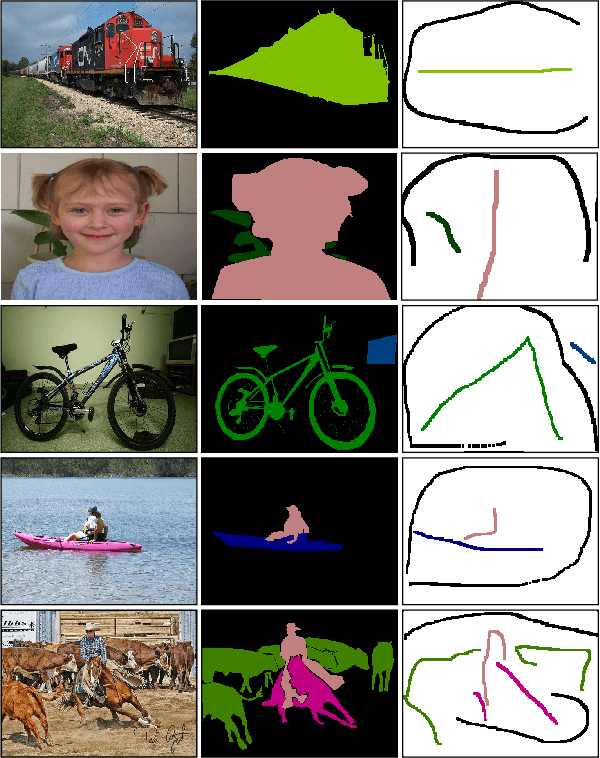
Abstract:A critical challenge of training deep learning models in the Digital Pathology (DP) domain is the high annotation cost by medical experts. One way to tackle this issue is via transfer learning from the natural image domain (NI), where the annotation cost is considerably cheaper. Cross-domain transfer learning from NI to DP is shown to be successful via class labels. One potential weakness of relying on class labels is the lack of spatial information, which can be obtained from spatial labels such as full pixel-wise segmentation labels and scribble labels. We demonstrate that scribble labels from NI domain can boost the performance of DP models on two cancer classification datasets (Patch Camelyon Breast Cancer and Colorectal Cancer dataset). Furthermore, we show that models trained with scribble labels yield the same performance boost as full pixel-wise segmentation labels despite being significantly easier and faster to collect.
The GIST and RIST of Iterative Self-Training for Semi-Supervised Segmentation
Mar 31, 2021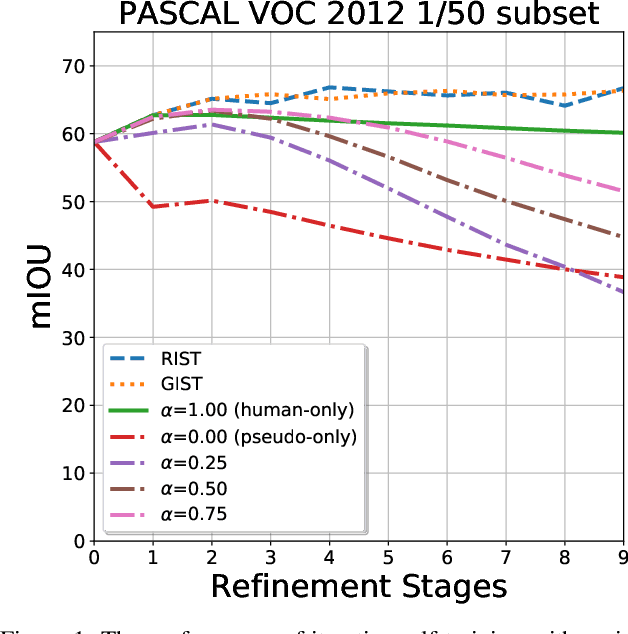
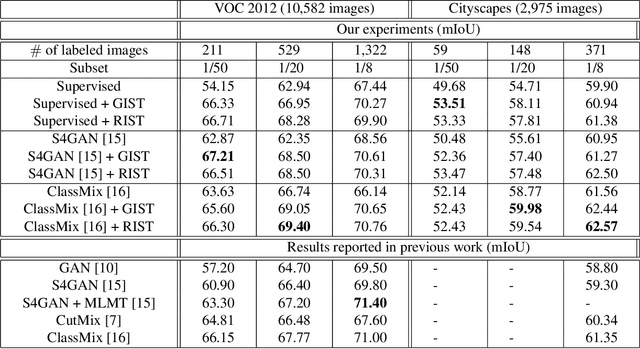
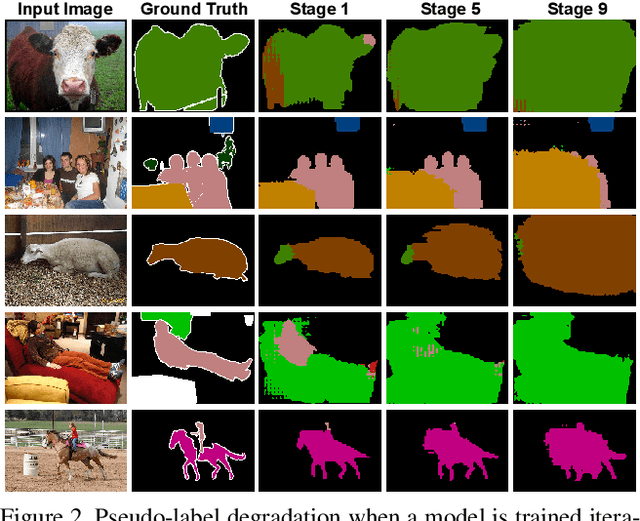
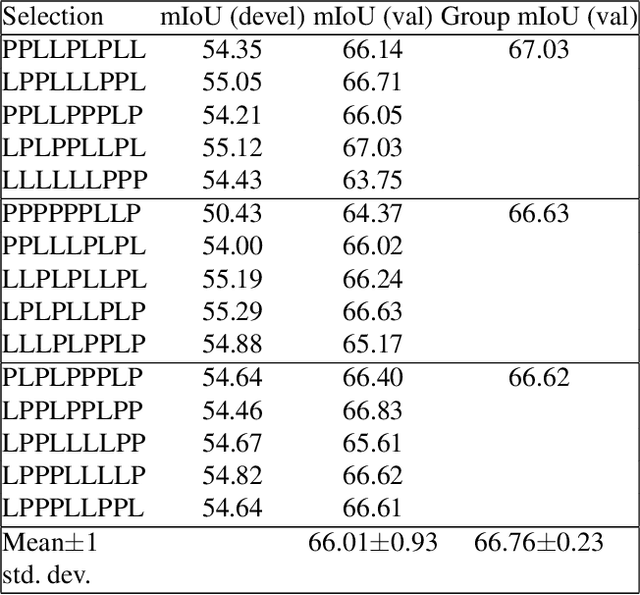
Abstract:We consider the task of semi-supervised semantic segmentation, where we aim to produce pixel-wise semantic object masks given only a small number of human-labeled training examples. We focus on iterative self-training methods in which we explore the behavior of self-training over multiple refinement stages. We show that iterative self-training leads to performance degradation if done naively with a fixed ratio of human-labeled to pseudo-labeled training examples. We propose Greedy Iterative Self-Training (GIST) and Random Iterative Self-Training (RIST) strategies that alternate between training on either human-labeled data or pseudo-labeled data at each refinement stage, resulting in a performance boost rather than degradation. We further show that GIST and RIST can be combined with existing SOTA methods to boost performance, yielding new SOTA results in Pascal VOC 2012 and Cityscapes dataset across five out of six subsets.
ProxyNCA++: Revisiting and Revitalizing Proxy Neighborhood Component Analysis
Apr 02, 2020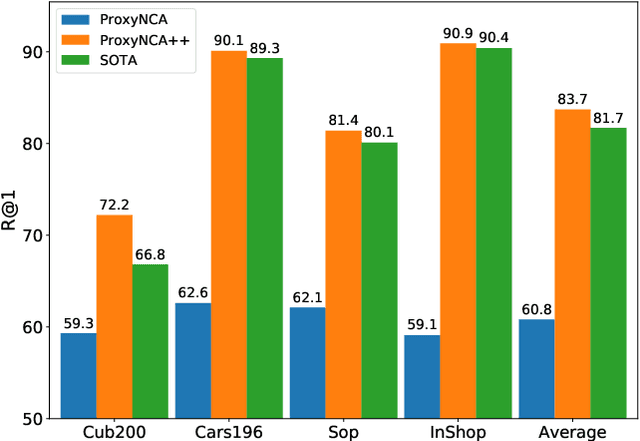
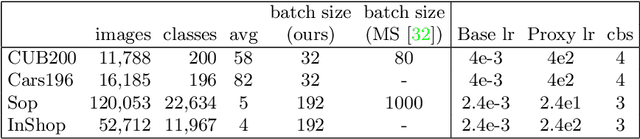
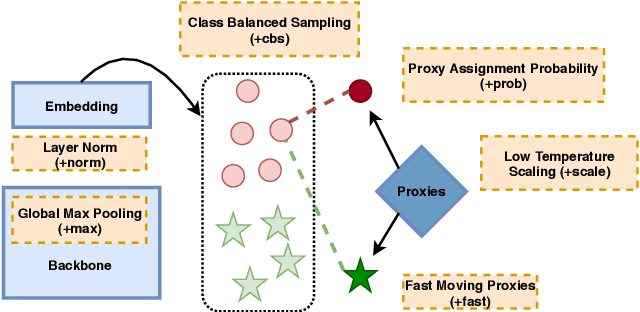
Abstract:We consider the problem of distance metric learning (DML), where the task is to learn an effective similarity measure between images. We revisit ProxyNCA and incorporate several enhancements. We find that low temperature scaling is a performance-critical component and explain why it works. Besides, we also discover that Global Max Pooling works better in general when compared to Global Average Pooling. Additionally, our proposed fast moving proxies also addresses small gradient issue of proxies, and this component synergizes well with low temperature scaling and Global Max Pooling. Our enhanced model, called ProxyNCA++, achieves a 22.9 percentage point average improvement of Recall@1 across four different zero-shot retrieval datasets compared to the original ProxyNCA algorithm. Furthermore, we achieve state-of-the-art results on the CUB200, Cars196, Sop, and InShop datasets, achieving Recall@1 scores of 72.2, 90.1, 81.4, and 90.9, respectively.
Learning with less data via Weakly Labeled Patch Classification in Digital Pathology
Jan 10, 2020
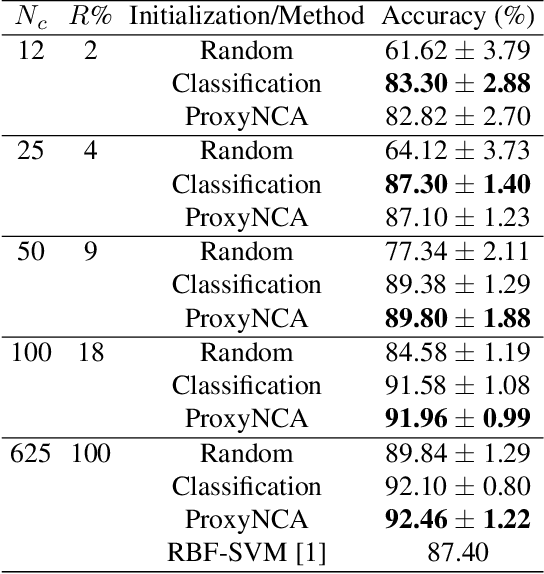
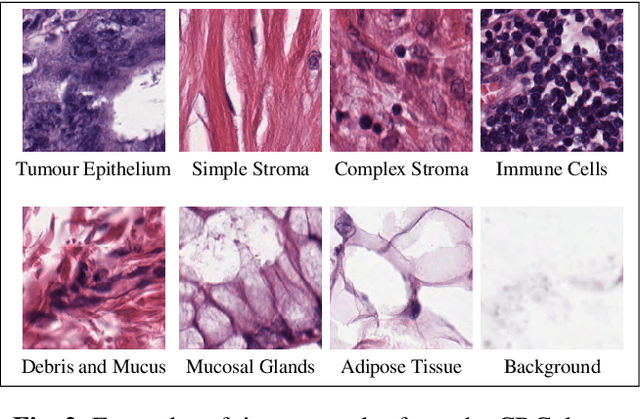
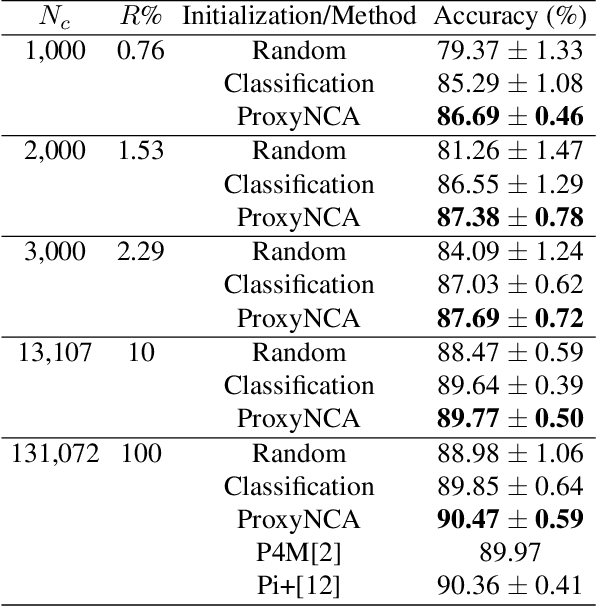
Abstract:In Digital Pathology (DP), labeled data is generally very scarce due to the requirement that medical experts provide annotations. We address this issue by learning transferable features from weakly labeled data, which are collected from various parts of the body and are organized by non-medical experts. In this paper, we show that features learned from such weakly labeled datasets are indeed transferable and allow us to achieve highly competitive patch classification results on the colorectal cancer (CRC) dataset [1] and the PatchCamelyon (PCam) dataset [2] while using an order of magnitude less labeled data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge