Enze Ye
Pose-Free 3D Quantitative Phase Imaging of Flowing Cellular Populations
Sep 05, 2025Abstract:High-throughput 3D quantitative phase imaging (QPI) in flow cytometry enables label-free, volumetric characterization of individual cells by reconstructing their refractive index (RI) distributions from multiple viewing angles during flow through microfluidic channels. However, current imaging methods assume that cells undergo uniform, single-axis rotation, which require their poses to be known at each frame. This assumption restricts applicability to near-spherical cells and prevents accurate imaging of irregularly shaped cells with complex rotations. As a result, only a subset of the cellular population can be analyzed, limiting the ability of flow-based assays to perform robust statistical analysis. We introduce OmniFHT, a pose-free 3D RI reconstruction framework that leverages the Fourier diffraction theorem and implicit neural representations (INRs) for high-throughput flow cytometry tomographic imaging. By jointly optimizing each cell's unknown rotational trajectory and volumetric structure under weak scattering assumptions, OmniFHT supports arbitrary cell geometries and multi-axis rotations. Its continuous representation also allows accurate reconstruction from sparsely sampled projections and restricted angular coverage, producing high-fidelity results with as few as 10 views or only 120 degrees of angular range. OmniFHT enables, for the first time, in situ, high-throughput tomographic imaging of entire flowing cell populations, providing a scalable and unbiased solution for label-free morphometric analysis in flow cytometry platforms.
Learning Diffusion Model from Noisy Measurement using Principled Expectation-Maximization Method
Oct 15, 2024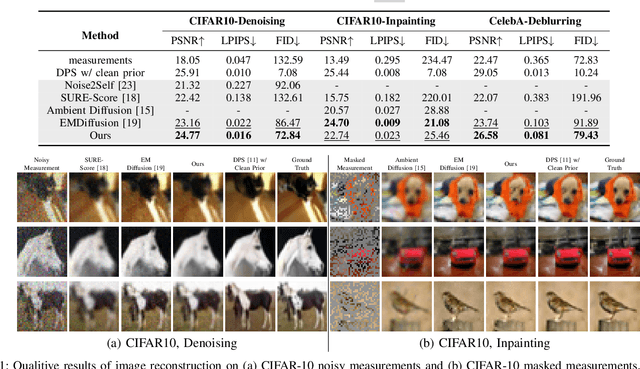

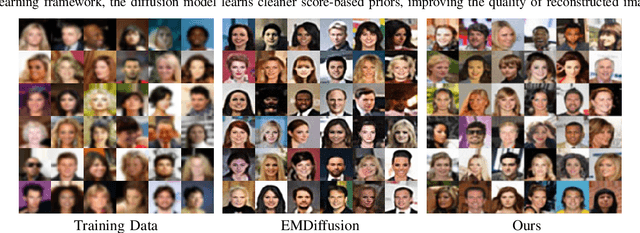
Abstract:Diffusion models have demonstrated exceptional ability in modeling complex image distributions, making them versatile plug-and-play priors for solving imaging inverse problems. However, their reliance on large-scale clean datasets for training limits their applicability in scenarios where acquiring clean data is costly or impractical. Recent approaches have attempted to learn diffusion models directly from corrupted measurements, but these methods either lack theoretical convergence guarantees or are restricted to specific types of data corruption. In this paper, we propose a principled expectation-maximization (EM) framework that iteratively learns diffusion models from noisy data with arbitrary corruption types. Our framework employs a plug-and-play Monte Carlo method to accurately estimate clean images from noisy measurements, followed by training the diffusion model using the reconstructed images. This process alternates between estimation and training until convergence. We evaluate the performance of our method across various imaging tasks, including inpainting, denoising, and deblurring. Experimental results demonstrate that our approach enables the learning of high-fidelity diffusion priors from noisy data, significantly enhancing reconstruction quality in imaging inverse problems.
Recovering a Molecule's 3D Dynamics from Liquid-phase Electron Microscopy Movies
Aug 23, 2023Abstract:The dynamics of biomolecules are crucial for our understanding of their functioning in living systems. However, current 3D imaging techniques, such as cryogenic electron microscopy (cryo-EM), require freezing the sample, which limits the observation of their conformational changes in real time. The innovative liquid-phase electron microscopy (liquid-phase EM) technique allows molecules to be placed in the native liquid environment, providing a unique opportunity to observe their dynamics. In this paper, we propose TEMPOR, a Temporal Electron MicroscoPy Object Reconstruction algorithm for liquid-phase EM that leverages an implicit neural representation (INR) and a dynamical variational auto-encoder (DVAE) to recover time series of molecular structures. We demonstrate its advantages in recovering different motion dynamics from two simulated datasets, 7bcq and Cas9. To our knowledge, our work is the first attempt to directly recover 3D structures of a temporally-varying particle from liquid-phase EM movies. It provides a promising new approach for studying molecules' 3D dynamics in structural biology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge