Enmin Song
YOLO-OB: An improved anchor-free real-time multiscale colon polyp detector in colonoscopy
Dec 14, 2023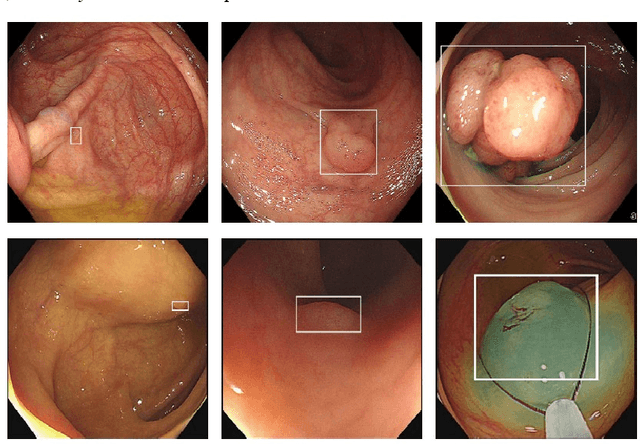



Abstract:Colon cancer is expected to become the second leading cause of cancer death in the United States in 2023. Although colonoscopy is one of the most effective methods for early prevention of colon cancer, up to 30% of polyps may be missed by endoscopists, thereby increasing patients' risk of developing colon cancer. Though deep neural networks have been proven to be an effective means of enhancing the detection rate of polyps. However, the variation of polyp size brings the following problems: (1) it is difficult to design an efficient and sufficient multi-scale feature fusion structure; (2) matching polyps of different sizes with fixed-size anchor boxes is a hard challenge. These problems reduce the performance of polyp detection and also lower the model's training and detection efficiency. To address these challenges, this paper proposes a new model called YOLO-OB. Specifically, we developed a bidirectional multiscale feature fusion structure, BiSPFPN, which could enhance the feature fusion capability across different depths of a CNN. We employed the ObjectBox detection head, which used a center-based anchor-free box regression strategy that could detect polyps of different sizes on feature maps of any scale. Experiments on the public dataset SUN and the self-collected colon polyp dataset Union demonstrated that the proposed model significantly improved various performance metrics of polyp detection, especially the recall rate. Compared to the state-of-the-art results on the public dataset SUN, the proposed method achieved a 6.73% increase on recall rate from 91.5% to 98.23%. Furthermore, our YOLO-OB was able to achieve real-time polyp detection at a speed of 39 frames per second using a RTX3090 graphics card. The implementation of this paper can be found here: https://github.com/seanyan62/YOLO-OB.
An Unpaired Cross-modality Segmentation Framework Using Data Augmentation and Hybrid Convolutional Networks for Segmenting Vestibular Schwannoma and Cochlea
Nov 28, 2022Abstract:The crossMoDA challenge aims to automatically segment the vestibular schwannoma (VS) tumor and cochlea regions of unlabeled high-resolution T2 scans by leveraging labeled contrast-enhanced T1 scans. The 2022 edition extends the segmentation task by including multi-institutional scans. In this work, we proposed an unpaired cross-modality segmentation framework using data augmentation and hybrid convolutional networks. Considering heterogeneous distributions and various image sizes for multi-institutional scans, we apply the min-max normalization for scaling the intensities of all scans between -1 and 1, and use the voxel size resampling and center cropping to obtain fixed-size sub-volumes for training. We adopt two data augmentation methods for effectively learning the semantic information and generating realistic target domain scans: generative and online data augmentation. For generative data augmentation, we use CUT and CycleGAN to generate two groups of realistic T2 volumes with different details and appearances for supervised segmentation training. For online data augmentation, we design a random tumor signal reducing method for simulating the heterogeneity of VS tumor signals. Furthermore, we utilize an advanced hybrid convolutional network with multi-dimensional convolutions to adaptively learn sparse inter-slice information and dense intra-slice information for accurate volumetric segmentation of VS tumor and cochlea regions in anisotropic scans. On the crossMoDA2022 validation dataset, our method produces promising results and achieves the mean DSC values of 72.47% and 76.48% and ASSD values of 3.42 mm and 0.53 mm for VS tumor and cochlea regions, respectively.
A novel algorithm for segmentation of leukocytes in peripheral blood
May 21, 2019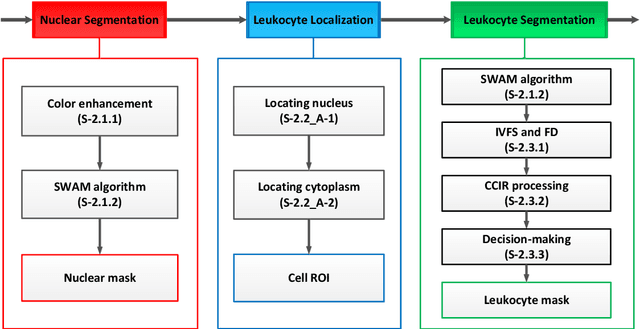
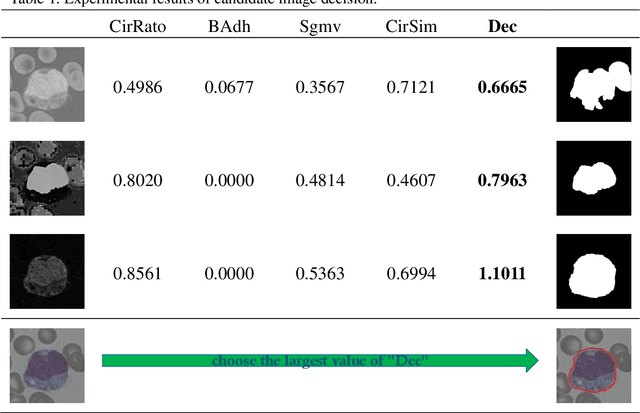
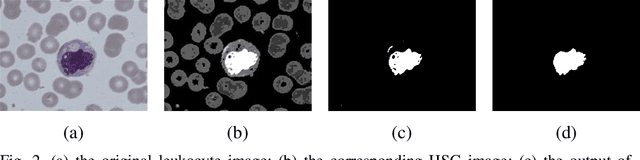

Abstract:In the detection of anemia, leukemia and other blood diseases, the number and type of leukocytes are essential evaluation parameters. However, the conventional leukocyte counting method is not only quite time-consuming but also error-prone. Consequently, many automation methods are introduced for the diagnosis of medical images. It remains difficult to accurately extract related features and count the number of cells under the variable conditions such as background, staining method, staining degree, light conditions and so on. Therefore, in order to adapt to various complex situations, we consider RGB color space, HSI color space, and the linear combination of G, H and S components, and propose a fast and accurate algorithm for the segmentation of peripheral blood leukocytes in this paper. First, the nucleus of leukocyte was separated by using the stepwise averaging method. Then based on the interval-valued fuzzy sets, the cytoplasm of leukocyte was segmented by minimizing the fuzzy divergence. Next, post-processing was carried out by using the concave-convex iterative repair algorithm and the decision mechanism of candidate mask sets. Experimental results show that the proposed method outperforms the existing non-fuzzy sets methods. Among the methods based on fuzzy sets, the interval-valued fuzzy sets perform slightly better than interval-valued intuitionistic fuzzy sets and intuitionistic fuzzy sets.
* 25 pages, 10 figures
Dual-branch residual network for lung nodule segmentation
May 21, 2019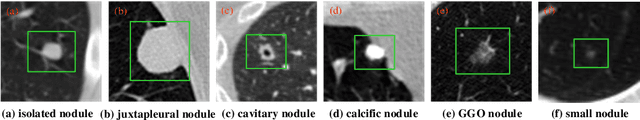
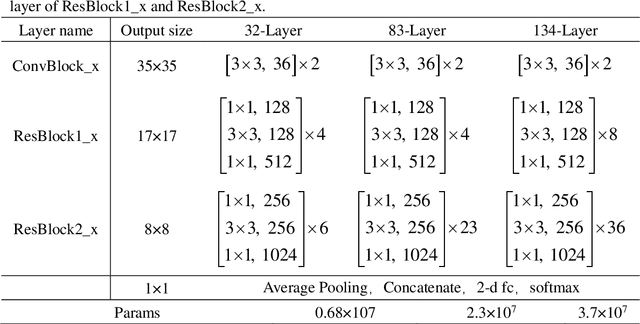
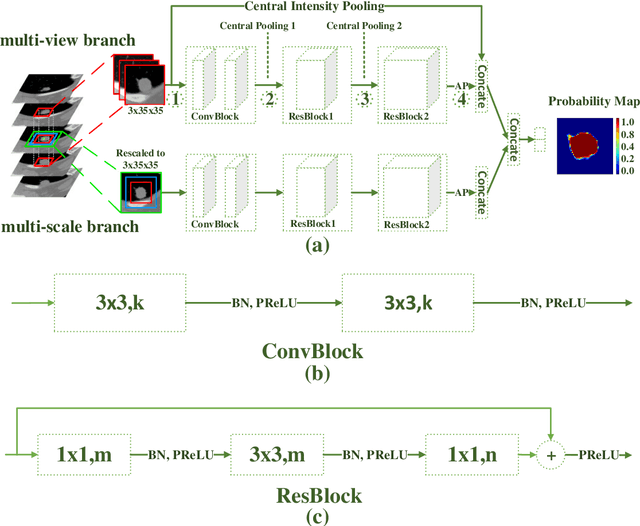
Abstract:An accurate segmentation of lung nodules in computed tomography (CT) images is critical to lung cancer analysis and diagnosis. However, due to the variety of lung nodules and the similarity of visual characteristics between nodules and their surroundings, a robust segmentation of nodules becomes a challenging problem. In this study, we propose the Dual-branch Residual Network (DB-ResNet) which is a data-driven model. Our approach integrates two new schemes to improve the generalization capability of the model: 1) the proposed model can simultaneously capture multi-view and multi-scale features of different nodules in CT images; 2) we combine the features of the intensity and the convolution neural networks (CNN). We propose a pooling method, called the central intensity-pooling layer (CIP), to extract the intensity features of the center voxel of the block, and then use the CNN to obtain the convolutional features of the center voxel of the block. In addition, we designed a weighted sampling strategy based on the boundary of nodules for the selection of those voxels using the weighting score, to increase the accuracy of the model. The proposed method has been extensively evaluated on the LIDC dataset containing 986 nodules. Experimental results show that the DB-ResNet achieves superior segmentation performance with an average dice score of 82.74% on the dataset. Moreover, we compared our results with those of four radiologists on the same dataset. The comparison showed that our average dice score was 0.49% higher than that of human experts. This proves that our proposed method is as good as the experienced radiologist.
Two-Stage Convolutional Neural Network Architecture for Lung Nodule Detection
May 09, 2019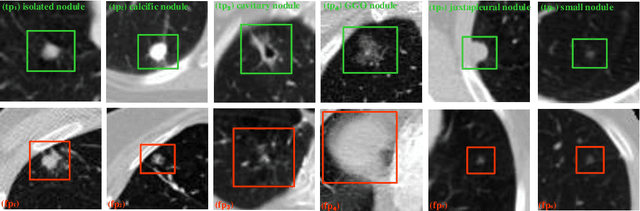
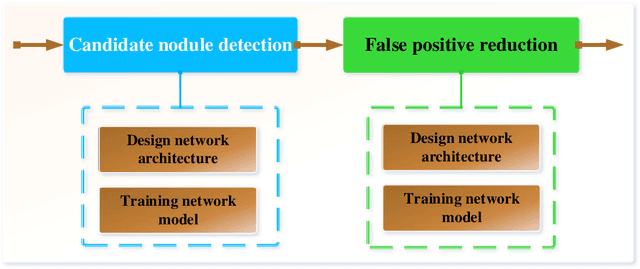

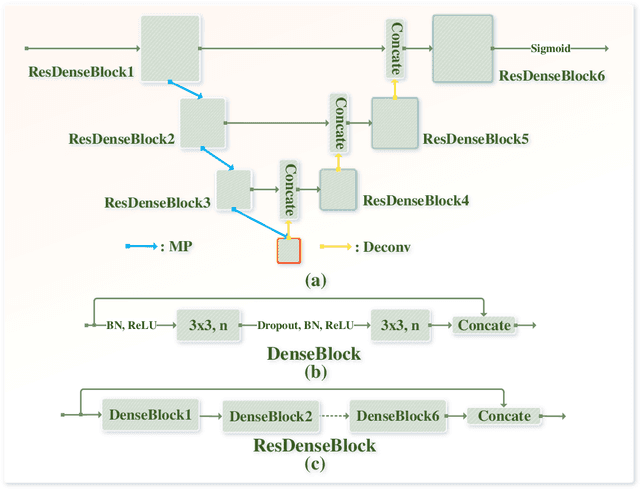
Abstract:Early detection of lung cancer is an effective way to improve the survival rate of patients. It is a critical step to have accurate detection of lung nodules in computed tomography (CT) images for the diagnosis of lung cancer. However, due to the heterogeneity of the lung nodules and the complexity of the surrounding environment, robust nodule detection has been a challenging task. In this study, we propose a two-stage convolutional neural network (TSCNN) architecture for lung nodule detection. The CNN architecture in the first stage is based on the improved UNet segmentation network to establish an initial detection of lung nodules. Simultaneously, in order to obtain a high recall rate without introducing excessive false positive nodules, we propose a novel sampling strategy, and use the offline hard mining idea for training and prediction according to the proposed cascaded prediction method. The CNN architecture in the second stage is based on the proposed dual pooling structure, which is built into three 3D CNN classification networks for false positive reduction. Since the network training requires a significant amount of training data, we adopt a data augmentation method based on random mask. Furthermore, we have improved the generalization ability of the false positive reduction model by means of ensemble learning. The proposed method has been experimentally verified on the LUNA dataset. Experimental results show that the proposed TSCNN architecture can obtain competitive detection performance.
Bone marrow cells detection: A technique for the microscopic image analysis
May 05, 2018
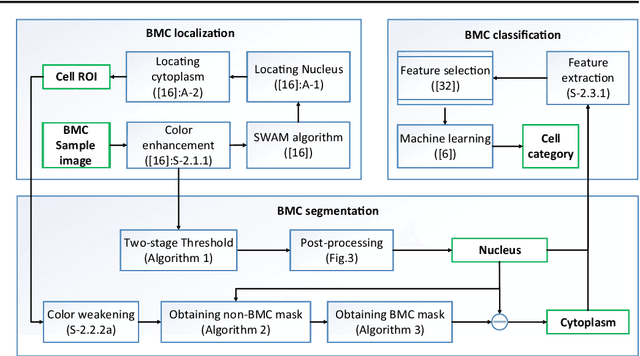
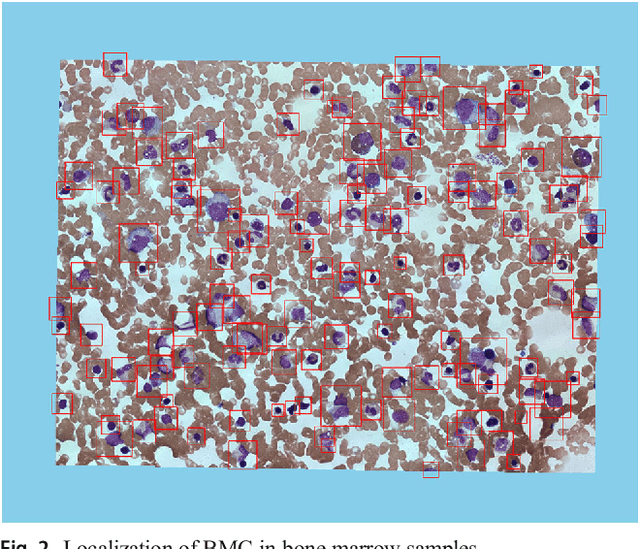
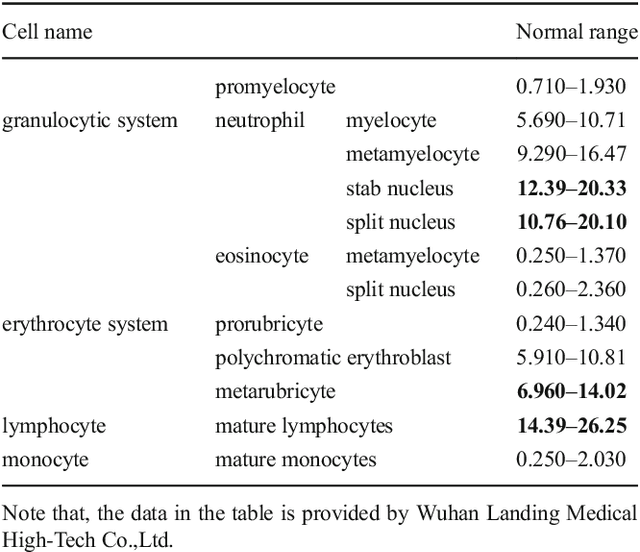
Abstract:In the detection of myeloproliferative, the number of cells in each type of bone marrow cells (BMC) is an important parameter for the evaluation. In this study, we propose a new counting method, which also consists of three modules including localization, segmentation and classification. The localization of BMC is achieved from a color transformation enhanced BMC sample image and stepwise averaging method (SAM). In the nucleus segmentation, both SAM and Otsu's method will be applied to obtain a weighted threshold for segmenting the patch into nucleus and non-nucleus. In the cytoplasm segmentation, a color weakening transformation, an improved region growing method and the K-Means algorithm are used. The connected cells with BMC will be separated by the marker-controlled watershed algorithm. The features will be extracted for the classification after the segmentation. In this study, the BMC are classified using the SVM, Random Forest, Artificial Neural Networks, Adaboost and Bayesian Networks into five classes including one outlier, namely, neutrophilic split granulocyte, neutrophilic stab granulocyte, metarubricyte, mature lymphocytes and the outlier (all other cells not listed). Our experimental results show that the best average recognition rate is 87.49% for the SVM.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge