Haichao Cao
Breast mass detection in digital mammography based on anchor-free architecture
Sep 02, 2020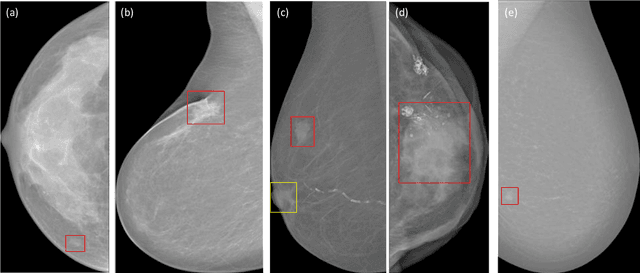


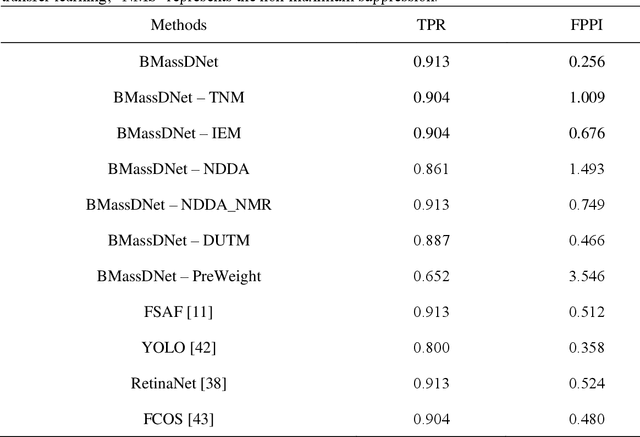
Abstract:Background and Objective: Accurate detection of breast masses in mammography images is critical to diagnose early breast cancer, which can greatly improve the patients survival rate. However, it is still a big challenge due to the heterogeneity of breast masses and the complexity of their surrounding environment.Methods: To address these problems, we propose a one-stage object detection architecture, called Breast Mass Detection Network (BMassDNet), based on anchor-free and feature pyramid which makes the detection of breast masses of different sizes well adapted. We introduce a truncation normalization method and combine it with adaptive histogram equalization to enhance the contrast between the breast mass and the surrounding environment. Meanwhile, to solve the overfitting problem caused by small data size, we propose a natural deformation data augmentation method and mend the train data dynamic updating method based on the data complexity to effectively utilize the limited data. Finally, we use transfer learning to assist the training process and to improve the robustness of the model ulteriorly.Results: On the INbreast dataset, each image has an average of 0.495 false positives whilst the recall rate is 0.930; On the DDSM dataset, when each image has 0.599 false positives, the recall rate reaches 0.943.Conclusions: The experimental results on datasets INbreast and DDSM show that the proposed BMassDNet can obtain competitive detection performance over the current top ranked methods.
A novel algorithm for segmentation of leukocytes in peripheral blood
May 21, 2019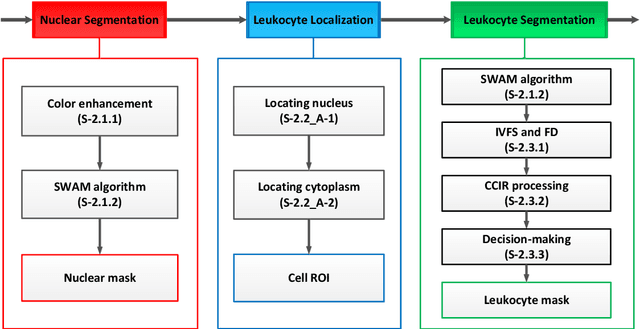
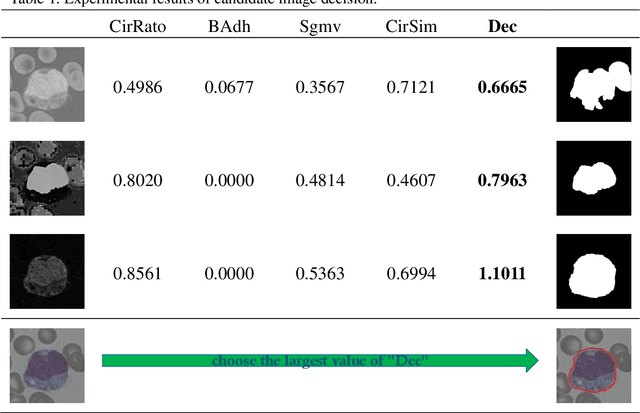
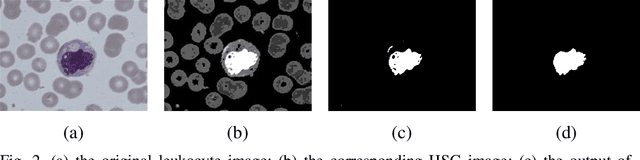

Abstract:In the detection of anemia, leukemia and other blood diseases, the number and type of leukocytes are essential evaluation parameters. However, the conventional leukocyte counting method is not only quite time-consuming but also error-prone. Consequently, many automation methods are introduced for the diagnosis of medical images. It remains difficult to accurately extract related features and count the number of cells under the variable conditions such as background, staining method, staining degree, light conditions and so on. Therefore, in order to adapt to various complex situations, we consider RGB color space, HSI color space, and the linear combination of G, H and S components, and propose a fast and accurate algorithm for the segmentation of peripheral blood leukocytes in this paper. First, the nucleus of leukocyte was separated by using the stepwise averaging method. Then based on the interval-valued fuzzy sets, the cytoplasm of leukocyte was segmented by minimizing the fuzzy divergence. Next, post-processing was carried out by using the concave-convex iterative repair algorithm and the decision mechanism of candidate mask sets. Experimental results show that the proposed method outperforms the existing non-fuzzy sets methods. Among the methods based on fuzzy sets, the interval-valued fuzzy sets perform slightly better than interval-valued intuitionistic fuzzy sets and intuitionistic fuzzy sets.
* 25 pages, 10 figures
Dual-branch residual network for lung nodule segmentation
May 21, 2019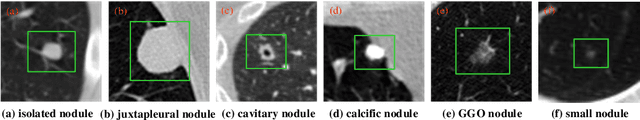
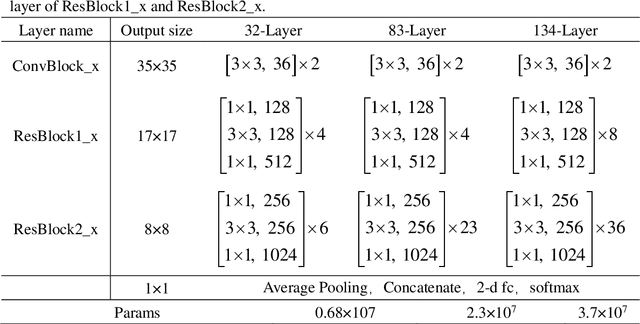
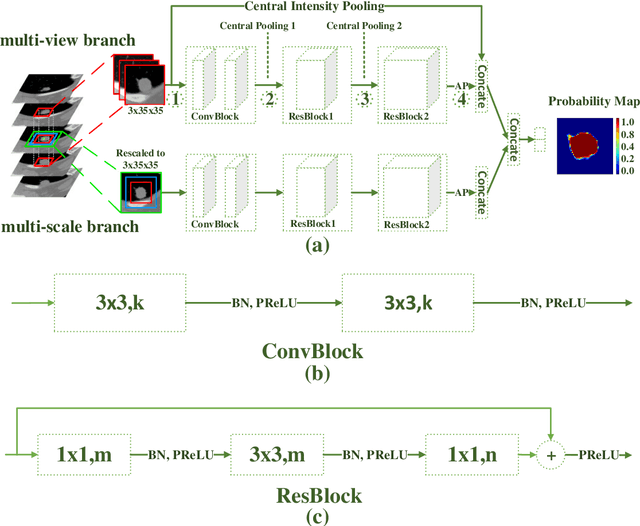
Abstract:An accurate segmentation of lung nodules in computed tomography (CT) images is critical to lung cancer analysis and diagnosis. However, due to the variety of lung nodules and the similarity of visual characteristics between nodules and their surroundings, a robust segmentation of nodules becomes a challenging problem. In this study, we propose the Dual-branch Residual Network (DB-ResNet) which is a data-driven model. Our approach integrates two new schemes to improve the generalization capability of the model: 1) the proposed model can simultaneously capture multi-view and multi-scale features of different nodules in CT images; 2) we combine the features of the intensity and the convolution neural networks (CNN). We propose a pooling method, called the central intensity-pooling layer (CIP), to extract the intensity features of the center voxel of the block, and then use the CNN to obtain the convolutional features of the center voxel of the block. In addition, we designed a weighted sampling strategy based on the boundary of nodules for the selection of those voxels using the weighting score, to increase the accuracy of the model. The proposed method has been extensively evaluated on the LIDC dataset containing 986 nodules. Experimental results show that the DB-ResNet achieves superior segmentation performance with an average dice score of 82.74% on the dataset. Moreover, we compared our results with those of four radiologists on the same dataset. The comparison showed that our average dice score was 0.49% higher than that of human experts. This proves that our proposed method is as good as the experienced radiologist.
Two-Stage Convolutional Neural Network Architecture for Lung Nodule Detection
May 09, 2019

Abstract:Early detection of lung cancer is an effective way to improve the survival rate of patients. It is a critical step to have accurate detection of lung nodules in computed tomography (CT) images for the diagnosis of lung cancer. However, due to the heterogeneity of the lung nodules and the complexity of the surrounding environment, robust nodule detection has been a challenging task. In this study, we propose a two-stage convolutional neural network (TSCNN) architecture for lung nodule detection. The CNN architecture in the first stage is based on the improved UNet segmentation network to establish an initial detection of lung nodules. Simultaneously, in order to obtain a high recall rate without introducing excessive false positive nodules, we propose a novel sampling strategy, and use the offline hard mining idea for training and prediction according to the proposed cascaded prediction method. The CNN architecture in the second stage is based on the proposed dual pooling structure, which is built into three 3D CNN classification networks for false positive reduction. Since the network training requires a significant amount of training data, we adopt a data augmentation method based on random mask. Furthermore, we have improved the generalization ability of the false positive reduction model by means of ensemble learning. The proposed method has been experimentally verified on the LUNA dataset. Experimental results show that the proposed TSCNN architecture can obtain competitive detection performance.
Bone marrow cells detection: A technique for the microscopic image analysis
May 05, 2018
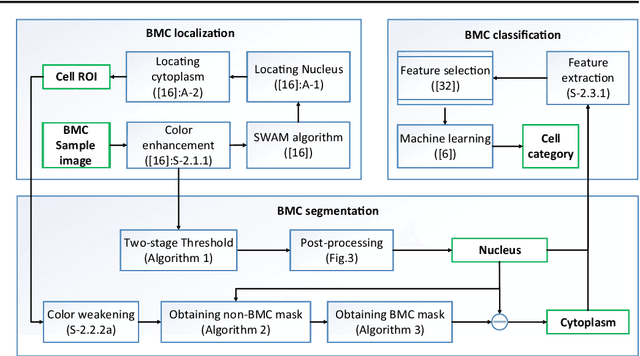
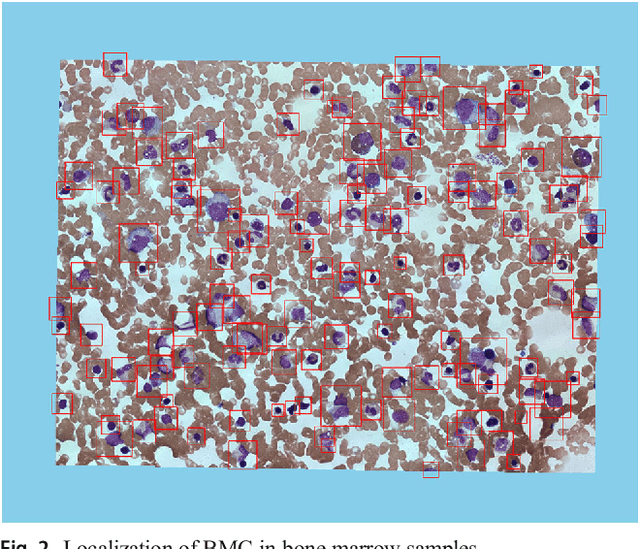
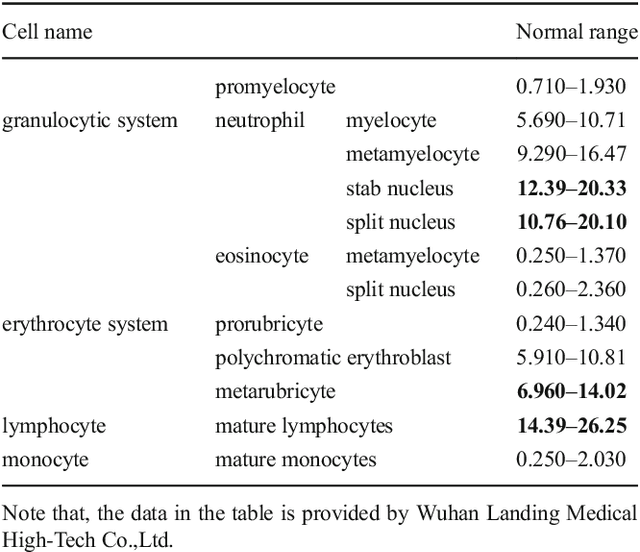
Abstract:In the detection of myeloproliferative, the number of cells in each type of bone marrow cells (BMC) is an important parameter for the evaluation. In this study, we propose a new counting method, which also consists of three modules including localization, segmentation and classification. The localization of BMC is achieved from a color transformation enhanced BMC sample image and stepwise averaging method (SAM). In the nucleus segmentation, both SAM and Otsu's method will be applied to obtain a weighted threshold for segmenting the patch into nucleus and non-nucleus. In the cytoplasm segmentation, a color weakening transformation, an improved region growing method and the K-Means algorithm are used. The connected cells with BMC will be separated by the marker-controlled watershed algorithm. The features will be extracted for the classification after the segmentation. In this study, the BMC are classified using the SVM, Random Forest, Artificial Neural Networks, Adaboost and Bayesian Networks into five classes including one outlier, namely, neutrophilic split granulocyte, neutrophilic stab granulocyte, metarubricyte, mature lymphocytes and the outlier (all other cells not listed). Our experimental results show that the best average recognition rate is 87.49% for the SVM.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge