Dongjin Choi
WellFactor: Patient Profiling using Integrative Embedding of Healthcare Data
Dec 21, 2023



Abstract:In the rapidly evolving healthcare industry, platforms now have access to not only traditional medical records, but also diverse data sets encompassing various patient interactions, such as those from healthcare web portals. To address this rich diversity of data, we introduce WellFactor: a method that derives patient profiles by integrating information from these sources. Central to our approach is the utilization of constrained low-rank approximation. WellFactor is optimized to handle the sparsity that is often inherent in healthcare data. Moreover, by incorporating task-specific label information, our method refines the embedding results, offering a more informed perspective on patients. One important feature of WellFactor is its ability to compute embeddings for new, previously unobserved patient data instantaneously, eliminating the need to revisit the entire data set or recomputing the embedding. Comprehensive evaluations on real-world healthcare data demonstrate WellFactor's effectiveness. It produces better results compared to other existing methods in classification performance, yields meaningful clustering of patients, and delivers consistent results in patient similarity searches and predictions.
Patient Clustering via Integrated Profiling of Clinical and Digital Data
Aug 22, 2023


Abstract:We introduce a novel profile-based patient clustering model designed for clinical data in healthcare. By utilizing a method grounded on constrained low-rank approximation, our model takes advantage of patients' clinical data and digital interaction data, including browsing and search, to construct patient profiles. As a result of the method, nonnegative embedding vectors are generated, serving as a low-dimensional representation of the patients. Our model was assessed using real-world patient data from a healthcare web portal, with a comprehensive evaluation approach which considered clustering and recommendation capabilities. In comparison to other baselines, our approach demonstrated superior performance in terms of clustering coherence and recommendation accuracy.
TagRuler: Interactive Tool for Span-Level Data Programming by Demonstration
Jun 24, 2021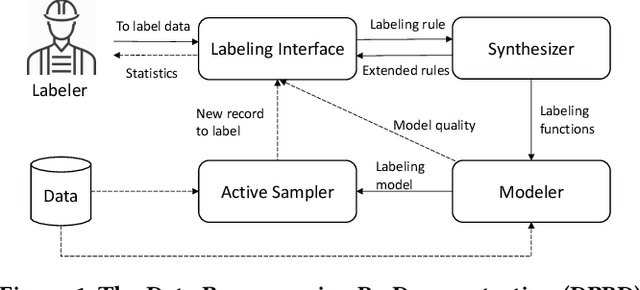
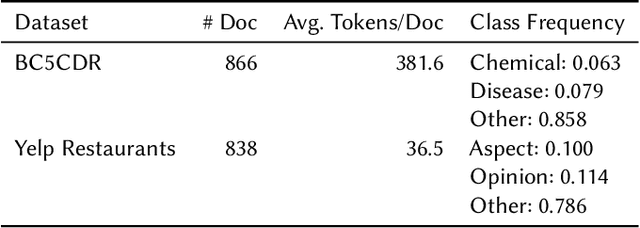
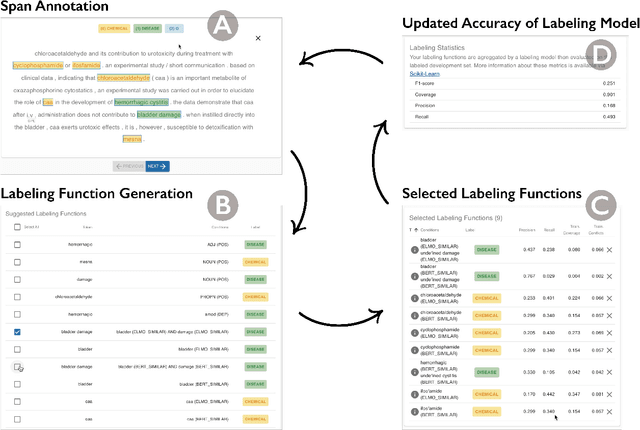
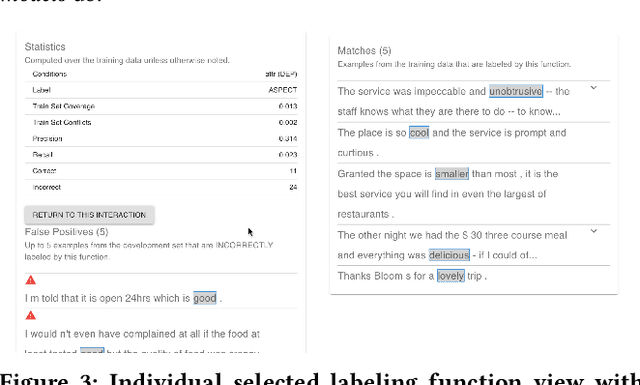
Abstract:Despite rapid developments in the field of machine learning research, collecting high-quality labels for supervised learning remains a bottleneck for many applications. This difficulty is exacerbated by the fact that state-of-the-art models for NLP tasks are becoming deeper and more complex, often increasing the amount of training data required even for fine-tuning. Weak supervision methods, including data programming, address this problem and reduce the cost of label collection by using noisy label sources for supervision. However, until recently, data programming was only accessible to users who knew how to program. To bridge this gap, the Data Programming by Demonstration framework was proposed to facilitate the automatic creation of labeling functions based on a few examples labeled by a domain expert. This framework has proven successful for generating high-accuracy labeling models for document classification. In this work, we extend the DPBD framework to span-level annotation tasks, arguably one of the most time-consuming NLP labeling tasks. We built a novel tool, TagRuler, that makes it easy for annotators to build span-level labeling functions without programming and encourages them to explore trade-offs between different labeling models and active learning strategies. We empirically demonstrated that an annotator could achieve a higher F1 score using the proposed tool compared to manual labeling for different span-level annotation tasks.
Putting Humans in the Natural Language Processing Loop: A Survey
Mar 06, 2021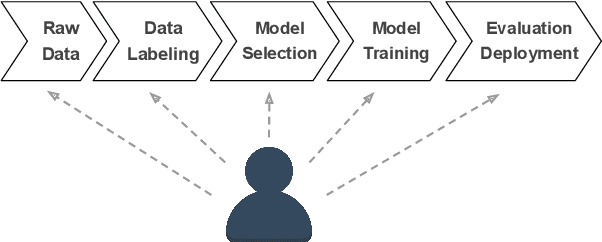
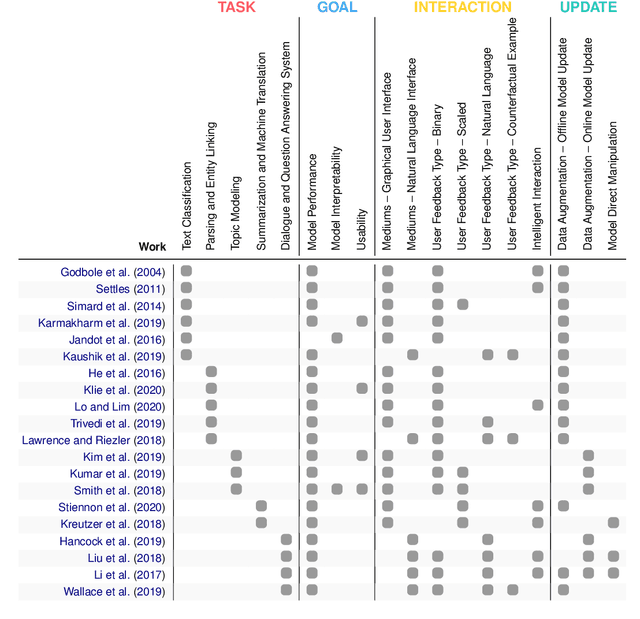
Abstract:How can we design Natural Language Processing (NLP) systems that learn from human feedback? There is a growing research body of Human-in-the-loop (HITL) NLP frameworks that continuously integrate human feedback to improve the model itself. HITL NLP research is nascent but multifarious -- solving various NLP problems, collecting diverse feedback from different people, and applying different methods to learn from collected feedback. We present a survey of HITL NLP work from both Machine Learning (ML) and Human-Computer Interaction (HCI) communities that highlights its short yet inspiring history, and thoroughly summarize recent frameworks focusing on their tasks, goals, human interactions, and feedback learning methods. Finally, we discuss future directions for integrating human feedback in the NLP development loop.
Data Programming by Demonstration: A Framework for Interactively Learning Labeling Functions
Sep 15, 2020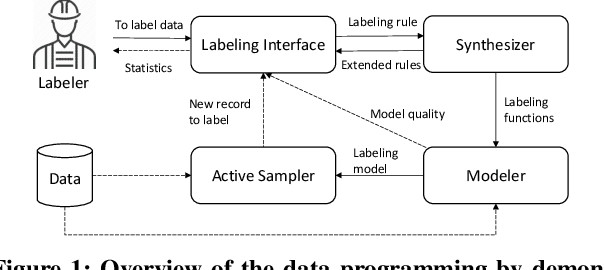
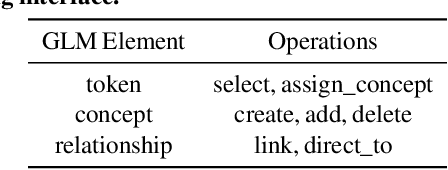
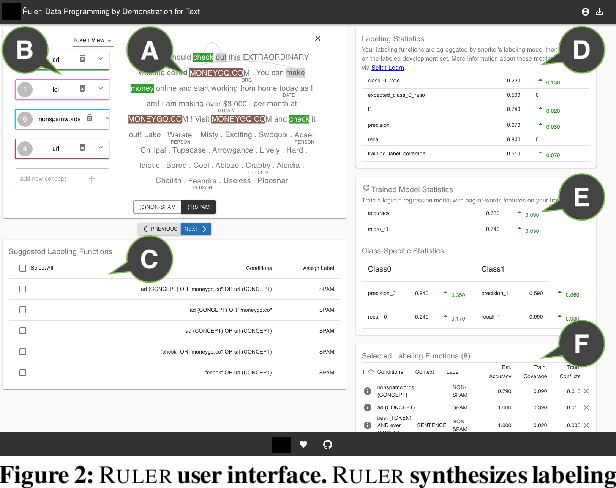

Abstract:Data programming is a programmatic weak supervision approach to efficiently curate large-scale labeled training data. Writing data programs (labeling functions) requires, however, both programming literacy and domain expertise. Many subject matter experts have neither programming proficiency nor time to effectively write data programs. Furthermore, regardless of one's expertise in coding or machine learning, transferring domain expertise into labeling functions by enumerating rules and thresholds is not only time consuming but also inherently difficult. Here we propose a new framework, data programming by demonstration (DPBD), to generate labeling rules using interactive demonstrations of users. DPBD aims to relieve the burden of writing labeling functions from users, enabling them to focus on higher-level semantics such as identifying relevant signals for labeling tasks. We operationalize our framework with Ruler, an interactive system that synthesizes labeling rules for document classification by using span-level annotations of users on document examples. We compare Ruler with conventional data programming through a user study conducted with 10 data scientists creating labeling functions for sentiment and spam classification tasks. We find that Ruler is easier to use and learn and offers higher overall satisfaction, while providing discriminative model performances comparable to ones achieved by conventional data programming.
SNeCT: Scalable network constrained Tucker decomposition for integrative multi-platform data analysis
Nov 26, 2017
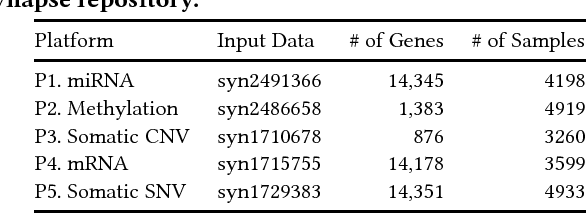
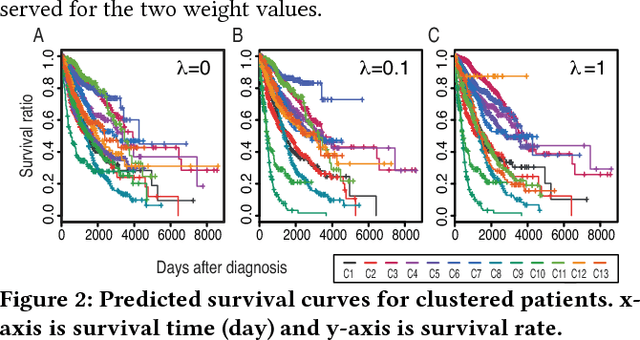
Abstract:Motivation: How do we integratively analyze large-scale multi-platform genomic data that are high dimensional and sparse? Furthermore, how can we incorporate prior knowledge, such as the association between genes, in the analysis systematically? Method: To solve this problem, we propose a Scalable Network Constrained Tucker decomposition method we call SNeCT. SNeCT adopts parallel stochastic gradient descent approach on the proposed parallelizable network constrained optimization function. SNeCT decomposition is applied to tensor constructed from large scale multi-platform multi-cohort cancer data, PanCan12, constrained on a network built from PathwayCommons database. Results: The decomposed factor matrices are applied to stratify cancers, to search for top-k similar patients, and to illustrate how the matrices can be used for personalized interpretation. In the stratification test, combined twelve-cohort data is clustered to form thirteen subclasses. The thirteen subclasses have a high correlation to tissue of origin in addition to other interesting observations, such as clear separation of OV cancers to two groups, and high clinical correlation within subclusters formed in cohorts BRCA and UCEC. In the top-k search, a new patient's genomic profile is generated and searched against existing patients based on the factor matrices. The similarity of the top-k patient to the query is high for 23 clinical features, including estrogen/progesterone receptor statuses of BRCA patients with average precision value ranges from 0.72 to 0.86 and from 0.68 to 0.86, respectively. We also provide an illustration of how the factor matrices can be used for interpretable personalized analysis of each patient.
CTD: Fast, Accurate, and Interpretable Method for Static and Dynamic Tensor Decompositions
Oct 09, 2017
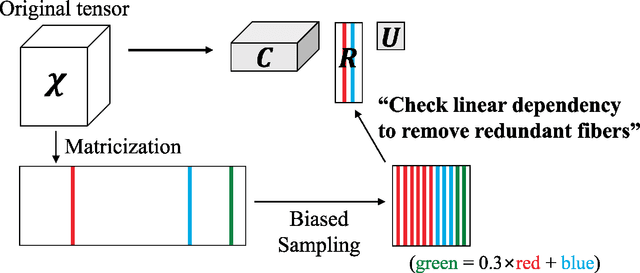

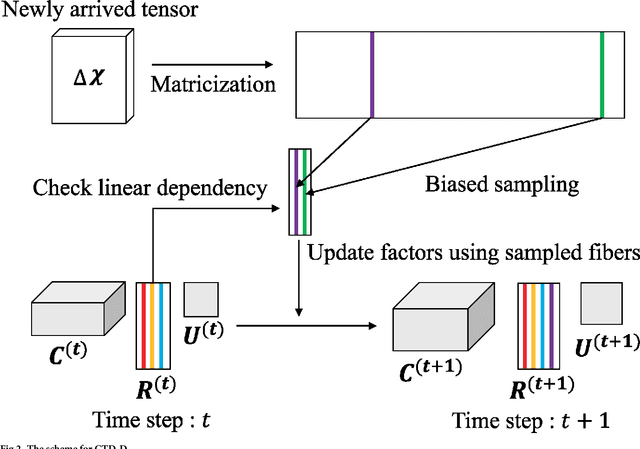
Abstract:How can we find patterns and anomalies in a tensor, or multi-dimensional array, in an efficient and directly interpretable way? How can we do this in an online environment, where a new tensor arrives each time step? Finding patterns and anomalies in a tensor is a crucial problem with many applications, including building safety monitoring, patient health monitoring, cyber security, terrorist detection, and fake user detection in social networks. Standard PARAFAC and Tucker decomposition results are not directly interpretable. Although a few sampling-based methods have previously been proposed towards better interpretability, they need to be made faster, more memory efficient, and more accurate. In this paper, we propose CTD, a fast, accurate, and directly interpretable tensor decomposition method based on sampling. CTD-S, the static version of CTD, provably guarantees a high accuracy that is 17 ~ 83x more accurate than that of the state-of-the-art method. Also, CTD-S is made 5 ~ 86x faster, and 7 ~ 12x more memory-efficient than the state-of-the-art method by removing redundancy. CTD-D, the dynamic version of CTD, is the first interpretable dynamic tensor decomposition method ever proposed. Also, it is made 2 ~ 3x faster than already fast CTD-S by exploiting factors at previous time step and by reordering operations. With CTD, we demonstrate how the results can be effectively interpreted in the online distributed denial of service (DDoS) attack detection.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge