Dong-Hyun Kim
Improving Noise Robust Audio-Visual Speech Recognition via Router-Gated Cross-Modal Feature Fusion
Aug 26, 2025Abstract:Robust audio-visual speech recognition (AVSR) in noisy environments remains challenging, as existing systems struggle to estimate audio reliability and dynamically adjust modality reliance. We propose router-gated cross-modal feature fusion, a novel AVSR framework that adaptively reweights audio and visual features based on token-level acoustic corruption scores. Using an audio-visual feature fusion-based router, our method down-weights unreliable audio tokens and reinforces visual cues through gated cross-attention in each decoder layer. This enables the model to pivot toward the visual modality when audio quality deteriorates. Experiments on LRS3 demonstrate that our approach achieves an 16.51-42.67% relative reduction in word error rate compared to AV-HuBERT. Ablation studies confirm that both the router and gating mechanism contribute to improved robustness under real-world acoustic noise.
Deformation-Aware Segmentation Network Robust to Motion Artifacts for Brain Tissue Segmentation using Disentanglement Learning
Dec 05, 2024Abstract:Motion artifacts caused by prolonged acquisition time are a significant challenge in Magnetic Resonance Imaging (MRI), hindering accurate tissue segmentation. These artifacts appear as blurred images that mimic tissue-like appearances, making segmentation difficult. This study proposes a novel deep learning framework that demonstrates superior performance in both motion correction and robust brain tissue segmentation in the presence of artifacts. The core concept lies in a complementary process: a disentanglement learning network progressively removes artifacts, leading to cleaner images and consequently, more accurate segmentation by a jointly trained motion estimation and segmentation network. This network generates three outputs: a motioncorrected image, a motion deformation map that identifies artifact-affected regions, and a brain tissue segmentation mask. This deformation serves as a guidance mechanism for the disentanglement process, aiding the model in recovering lost information or removing artificial structures introduced by the artifacts. Extensive in-vivo experiments on pediatric motion data demonstrate that our proposed framework outperforms state-of-the-art methods in segmenting motion-corrupted MRI scans.
* Medical Image Computing and Computer Assisted Intervention, MICCAI 2024
Toward Automated Detection of Microbleeds with Anatomical Scale Localization: A Complete Clinical Diagnosis Support Using Deep Learning
Jun 22, 2023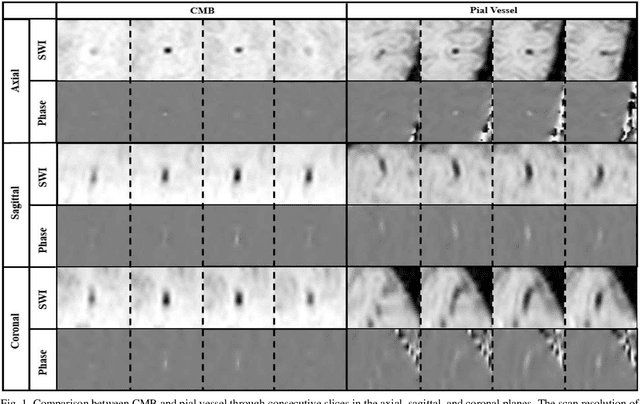
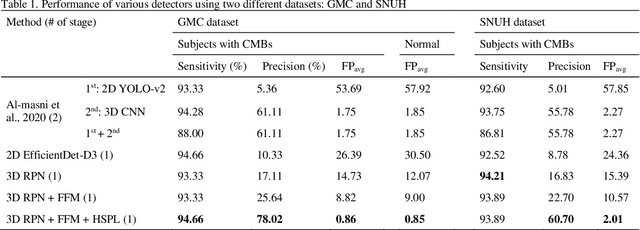
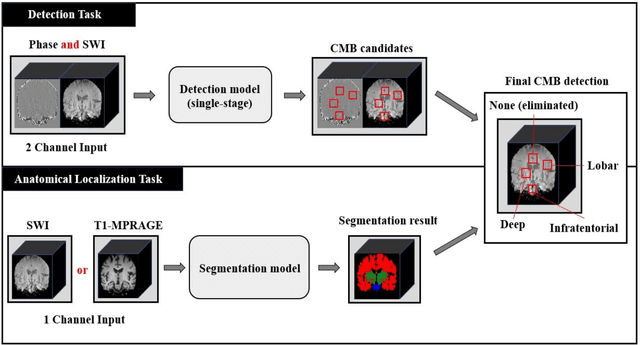
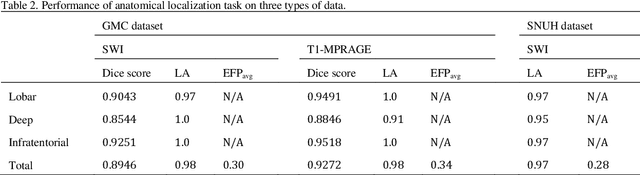
Abstract:Cerebral Microbleeds (CMBs) are chronic deposits of small blood products in the brain tissues, which have explicit relation to various cerebrovascular diseases depending on their anatomical location, including cognitive decline, intracerebral hemorrhage, and cerebral infarction. However, manual detection of CMBs is a time-consuming and error-prone process because of their sparse and tiny structural properties. The detection of CMBs is commonly affected by the presence of many CMB mimics that cause a high false-positive rate (FPR), such as calcification and pial vessels. This paper proposes a novel 3D deep learning framework that does not only detect CMBs but also inform their anatomical location in the brain (i.e., lobar, deep, and infratentorial regions). For the CMB detection task, we propose a single end-to-end model by leveraging the U-Net as a backbone with Region Proposal Network (RPN). To significantly reduce the FPs within the same single model, we develop a new scheme, containing Feature Fusion Module (FFM) that detects small candidates utilizing contextual information and Hard Sample Prototype Learning (HSPL) that mines CMB mimics and generates additional loss term called concentration loss using Convolutional Prototype Learning (CPL). The anatomical localization task does not only tell to which region the CMBs belong but also eliminate some FPs from the detection task by utilizing anatomical information. The results show that the proposed RPN that utilizes the FFM and HSPL outperforms the vanilla RPN and achieves a sensitivity of 94.66% vs. 93.33% and an average number of false positives per subject (FPavg) of 0.86 vs. 14.73. Also, the anatomical localization task further improves the detection performance by reducing the FPavg to 0.56 while maintaining the sensitivity of 94.66%.
Stacked U-Nets with Self-Assisted Priors Towards Robust Correction of Rigid Motion Artifact in Brain MRI
Nov 11, 2021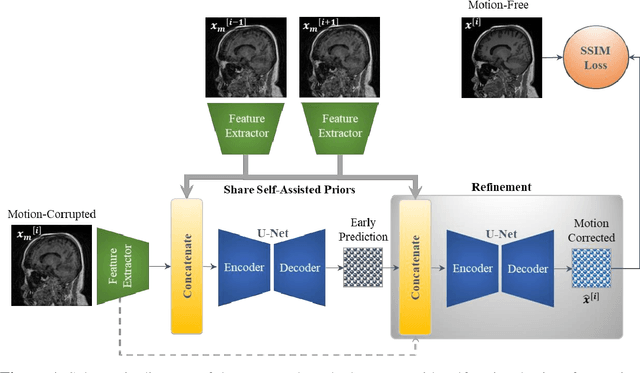

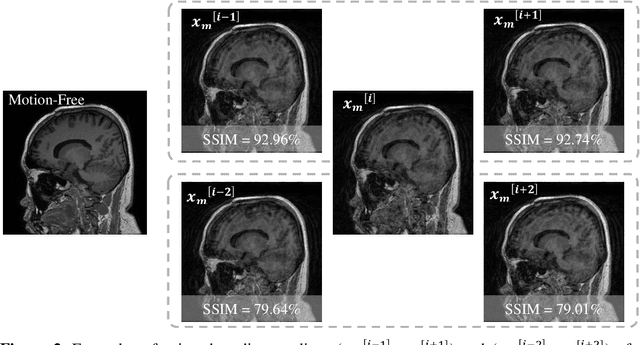
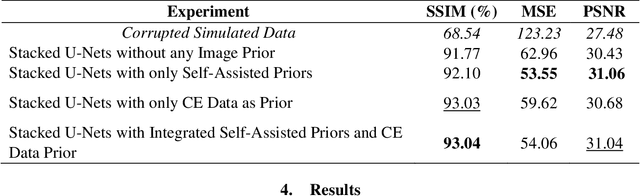
Abstract:In this paper, we develop an efficient retrospective deep learning method called stacked U-Nets with self-assisted priors to address the problem of rigid motion artifacts in MRI. The proposed work exploits the usage of additional knowledge priors from the corrupted images themselves without the need for additional contrast data. The proposed network learns missed structural details through sharing auxiliary information from the contiguous slices of the same distorted subject. We further design a refinement stacked U-Nets that facilitates preserving of the image spatial details and hence improves the pixel-to-pixel dependency. To perform network training, simulation of MRI motion artifacts is inevitable. We present an intensive analysis using various types of image priors: the proposed self-assisted priors and priors from other image contrast of the same subject. The experimental analysis proves the effectiveness and feasibility of our self-assisted priors since it does not require any further data scans.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge