David Gillespie
A Cloud-based Deep Learning Framework for Remote Detection of Diabetic Foot Ulcers
May 17, 2021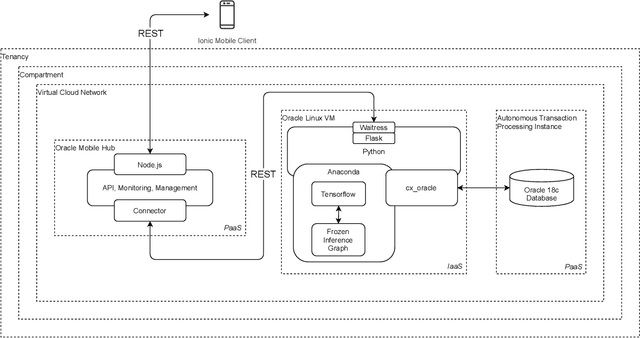

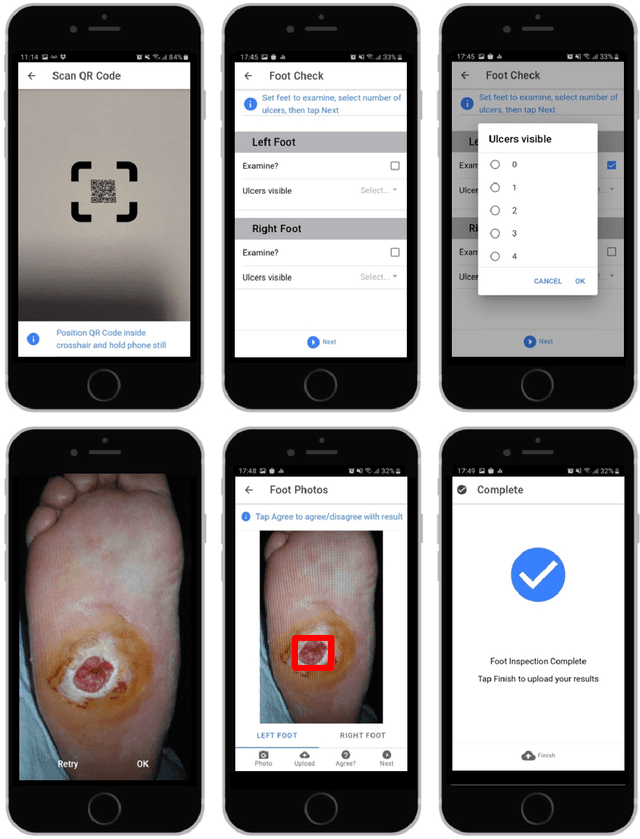
Abstract:This research proposes a mobile and cloud-based framework for the automatic detection of diabetic foot ulcers and conducts an investigation of its performance. The system uses a cross-platform mobile framework which enables the deployment of mobile apps to multiple platforms using a single TypeScript code base. A deep convolutional neural network was deployed to a cloud-based platform where the mobile app could send photographs of patient's feet for inference to detect the presence of diabetic foot ulcers. The functionality and usability of the system were tested in two clinical settings: Salford Royal NHS Foundation Trust and Lancashire Teaching Hospitals NHS Foundation Trust. The benefits of the system, such as the potential use of the app by patients to identify and monitor their condition are discussed.
Analysis Towards Classification of Infection and Ischaemia of Diabetic Foot Ulcers
Apr 07, 2021
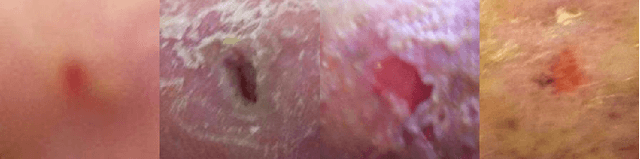
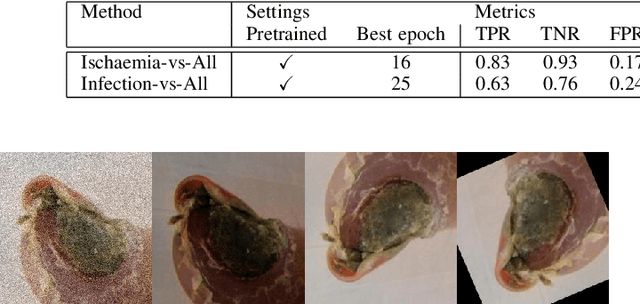
Abstract:This paper introduces the Diabetic Foot Ulcers dataset (DFUC2021) for analysis of pathology, focusing on infection and ischaemia. We describe the data preparation of DFUC2021 for ground truth annotation, data curation and data analysis. The final release of DFUC2021 consists of 15,683 DFU patches, with 5,955 training, 5,734 for testing and 3,994 unlabeled DFU patches. The ground truth labels are four classes, i.e. control, infection, ischaemia and both conditions. We curate the dataset using image hashing techniques and analyse the separability using UMAP projection. We benchmark the performance of five key backbones of deep learning, i.e. VGG16, ResNet101, InceptionV3, DenseNet121 and EfficientNet on DFUC2021. We report the optimised results of these key backbones with different strategies. Based on our observations, we conclude that EfficientNetB0 with data augmentation and transfer learning provided the best results for multi-class (4-class) classification with macro-average Precision, Recall and F1-score of 0.57, 0.62 and 0.55, respectively. In ischaemia and infection recognition, when trained on one-versus-all, EfficientNetB0 achieved comparable results with the state of the art. Finally, we interpret the results with statistical analysis and Grad-CAM visualisation.
Deep learning in magnetic resonance prostate segmentation: A review and a new perspective
Nov 16, 2020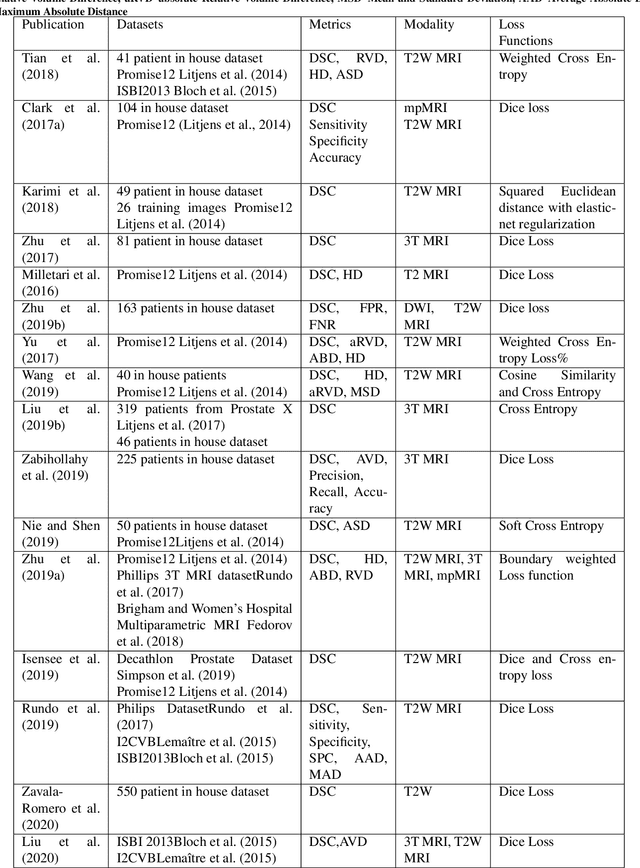
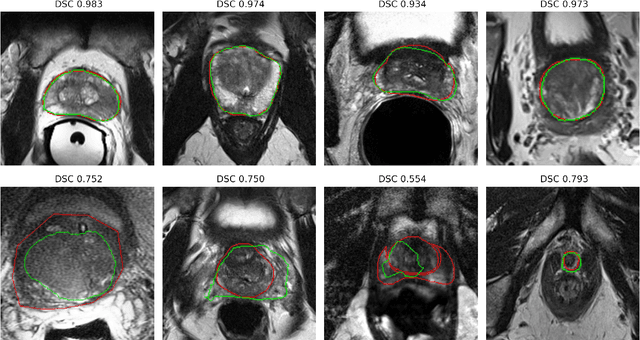
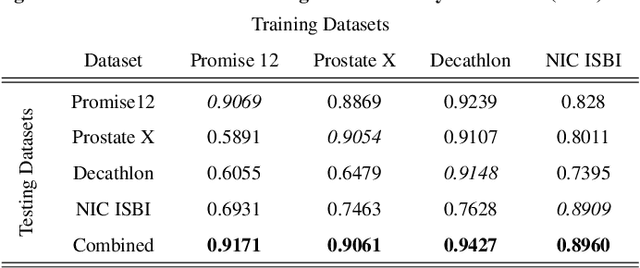
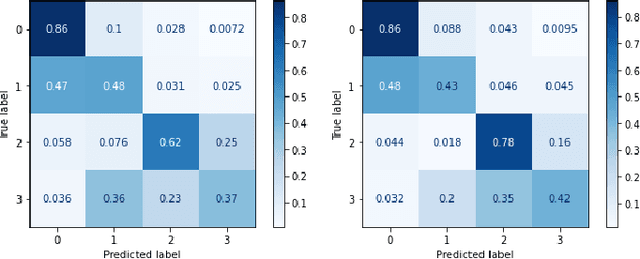
Abstract:Prostate radiotherapy is a well established curative oncology modality, which in future will use Magnetic Resonance Imaging (MRI)-based radiotherapy for daily adaptive radiotherapy target definition. However the time needed to delineate the prostate from MRI data accurately is a time consuming process. Deep learning has been identified as a potential new technology for the delivery of precision radiotherapy in prostate cancer, where accurate prostate segmentation helps in cancer detection and therapy. However, the trained models can be limited in their application to clinical setting due to different acquisition protocols, limited publicly available datasets, where the size of the datasets are relatively small. Therefore, to explore the field of prostate segmentation and to discover a generalisable solution, we review the state-of-the-art deep learning algorithms in MR prostate segmentation; provide insights to the field by discussing their limitations and strengths; and propose an optimised 2D U-Net for MR prostate segmentation. We evaluate the performance on four publicly available datasets using Dice Similarity Coefficient (DSC) as performance metric. Our experiments include within dataset evaluation and cross-dataset evaluation. The best result is achieved by composite evaluation (DSC of 0.9427 on Decathlon test set) and the poorest result is achieved by cross-dataset evaluation (DSC of 0.5892, Prostate X training set, Promise 12 testing set). We outline the challenges and provide recommendations for future work. Our research provides a new perspective to MR prostate segmentation and more importantly, we provide standardised experiment settings for researchers to evaluate their algorithms. Our code is available at https://github.com/AIEMMU/MRI\_Prostate.
Deep Learning in Diabetic Foot Ulcers Detection: A Comprehensive Evaluation
Oct 15, 2020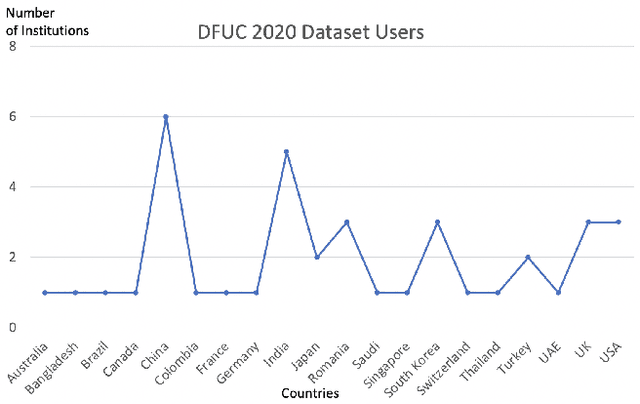
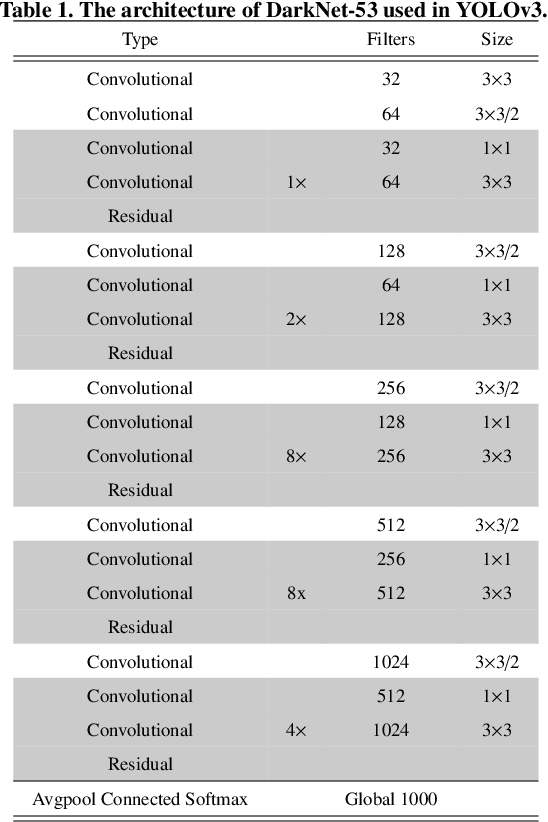
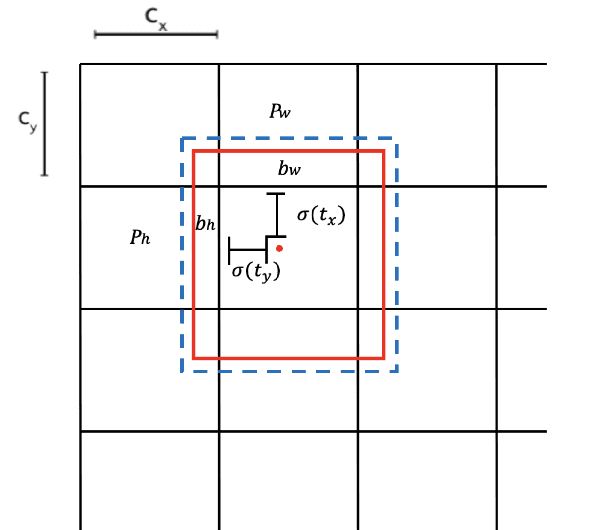
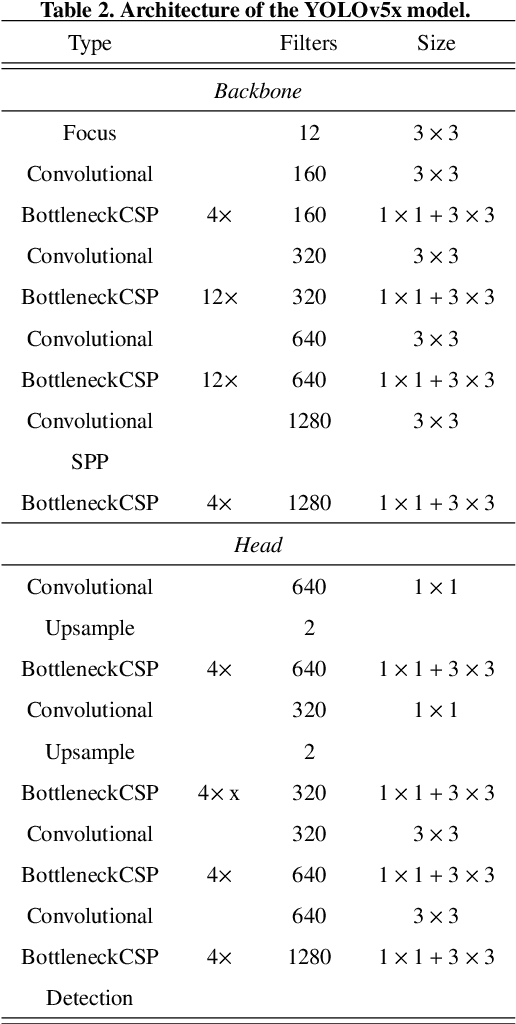
Abstract:There has been a substantial amount of research on computer methods and technology for the detection and recognition of diabetic foot ulcers (DFUs), but there is a lack of systematic comparisons of state-of-the-art deep learning object detection frameworks applied to this problem. With recent development and data sharing performed as part of the DFU Challenge (DFUC2020) such a comparison becomes possible: DFUC2020 provided participants with a comprehensive dataset consisting of 2,000 images for training each method and 2,000 images for testing them. The following deep learning-based algorithms are compared in this paper: Faster R-CNN, three variants of Faster R-CNN and an ensemble method; YOLOv3; YOLOv5; EfficientDet; and a new Cascade Attention Network. For each deep learning method, we provide a detailed description of model architecture, parameter settings for training and additional stages including pre-processing, data augmentation and post-processing. We provide a comprehensive evaluation for each method. All the methods required a data augmentation stage to increase the number of images available for training and a post-processing stage to remove false positives. The best performance is obtained Deformable Convolution, a variant of Faster R-CNN, with a mAP of 0.6940 and an F1-Score of 0.7434. Finally, we demonstrate that the ensemble method based on different deep learning methods can enhanced the F1-Score but not the mAP. Our results show that state-of-the-art deep learning methods can detect DFU with some accuracy, but there are many challenges ahead before they can be implemented in real world settings.
DFUC2020: Analysis Towards Diabetic Foot Ulcer Detection
Apr 24, 2020



Abstract:Every 20 seconds, a limb is amputated somewhere in the world due to diabetes. This is a global health problem that requires a global solution. The MICCAI challenge discussed in this paper, which concerns the detection of diabetic foot ulcers, will accelerate the development of innovative healthcare technology to address this unmet medical need. In an effort to improve patient care and reduce the strain on healthcare systems, recent research has focused on the creation of cloud-based detection algorithms that can be consumed as a service by a mobile app that patients (or a carer, partner or family member) could use themselves to monitor their condition and to detect the appearance of a diabetic foot ulcer (DFU). Collaborative work between Manchester Metropolitan University, Lancashire Teaching Hospital and the Manchester University NHS Foundation Trust has created a repository of 4000 DFU images for the purpose of supporting research toward more advanced methods of DFU detection. Based on a joint effort involving the lead scientists of the UK, US, India and New Zealand, this challenge will solicit original work, and promote interactions between researchers and interdisciplinary collaborations. This paper presents a dataset description and analysis, assessment methods, benchmark algorithms and initial evaluation results. It facilitates the challenge by providing useful insights into state-of-the-art and ongoing research.
Anysize GAN: A solution to the image-warping problem
Mar 06, 2020



Abstract:We propose a new type of General Adversarial Network (GAN) to resolve a common issue with Deep Learning. We develop a novel architecture that can be applied to existing latent vector based GAN structures that allows them to generate on-the-fly images of any size. Existing GAN for image generation requires uniform images of matching dimensions. However, publicly available datasets, such as ImageNet contain thousands of different sizes. Resizing image causes deformations and changing the image data, whereas as our network does not require this preprocessing step. We make significant changes to the standard data loading techniques to enable any size image to be loaded for training. We also modify the network in two ways, by adding multiple inputs and a novel dynamic resizing layer. Finally we make adjustments to the discriminator to work on multiple resolutions. These changes can allow multiple resolution datasets to be trained on without any resizing, if memory allows. We validate our results on the ISIC 2019 skin lesion dataset. We demonstrate our method can successfully generate realistic images at different sizes without issue, preserving and understanding spatial relationships, while maintaining feature relationships. We will release the source codes upon paper acceptance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge