David Epstein
Dense Steerable Filter CNNs for Exploiting Rotational Symmetry in Histology Images
Apr 06, 2020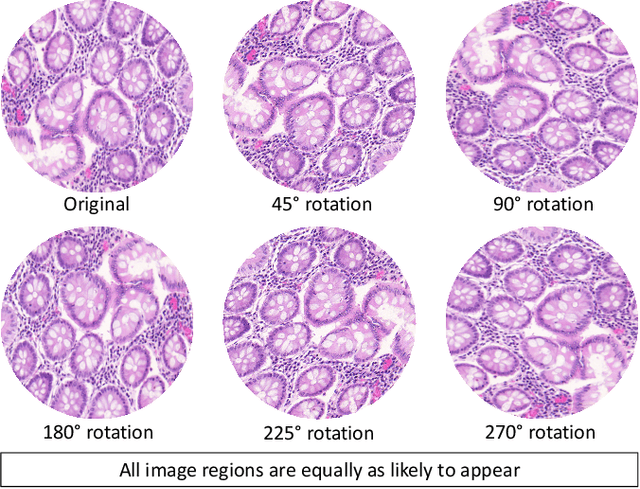
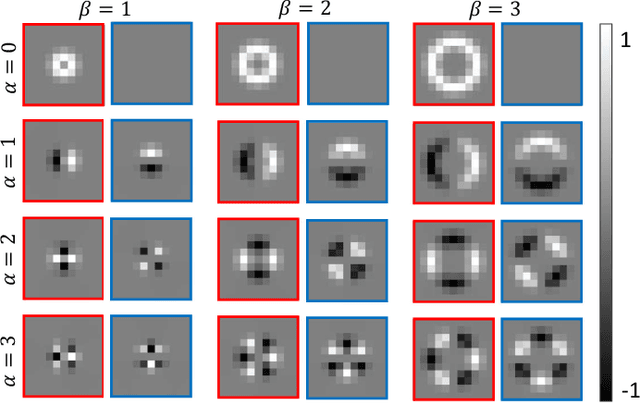
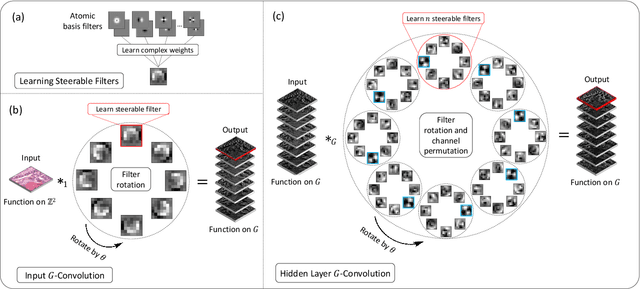
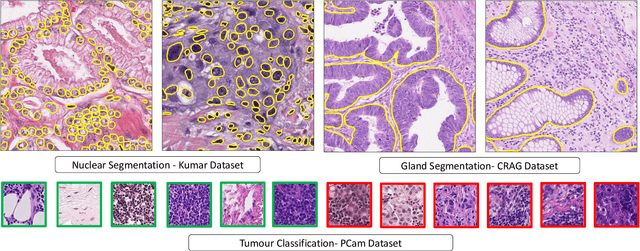
Abstract:Histology images are inherently symmetric under rotation, where each orientation is equally as likely to appear. However, this rotational symmetry is not widely utilised as prior knowledge in modern Convolutional Neural Networks (CNNs), resulting in data hungry models that learn independent features at each orientation. Allowing CNNs to be rotation-equivariant removes the necessity to learn this set of transformations from the data and instead frees up model capacity, allowing more discriminative features to be learned. This reduction in the number of required parameters also reduces the risk of overfitting. In this paper, we propose Dense Steerable Filter CNNs (DSF-CNNs) that use group convolutions with multiple rotated copies of each filter in a densely connected framework. Each filter is defined as a linear combination of steerable basis filters, enabling exact rotation and decreasing the number of trainable parameters compared to standard filters. We also provide the first in-depth comparison of different rotation-equivariant CNNs for histology image analysis and demonstrate the advantage of encoding rotational symmetry into modern architectures. We show that DSF-CNNs achieve state-of-the-art performance, with significantly fewer parameters, when applied to three different tasks in the area of computational pathology: breast tumour classification, colon gland segmentation and multi-tissue nuclear segmentation.
Fast and Accurate Tumor Segmentation of Histology Images using Persistent Homology and Deep Convolutional Features
May 09, 2018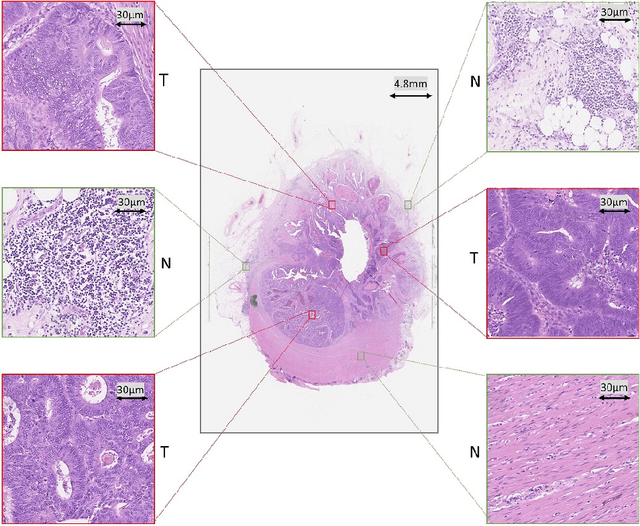
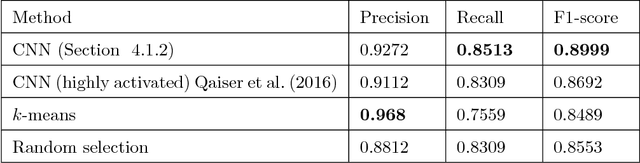
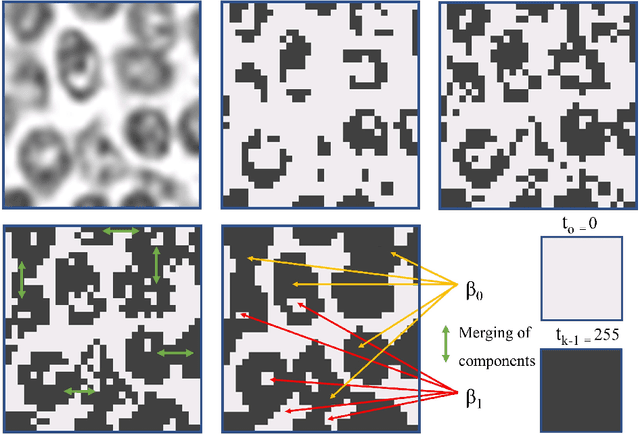
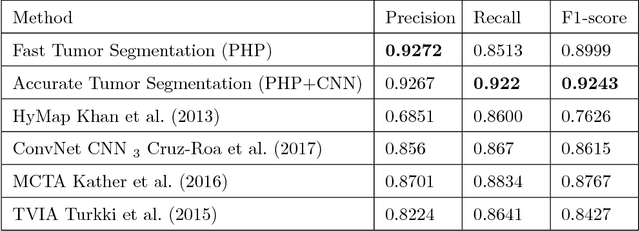
Abstract:Tumor segmentation in whole-slide images of histology slides is an important step towards computer-assisted diagnosis. In this work, we propose a tumor segmentation framework based on the novel concept of persistent homology profiles (PHPs). For a given image patch, the homology profiles are derived by efficient computation of persistent homology, which is an algebraic tool from homology theory. We propose an efficient way of computing topological persistence of an image, alternative to simplicial homology. The PHPs are devised to distinguish tumor regions from their normal counterparts by modeling the atypical characteristics of tumor nuclei. We propose two variants of our method for tumor segmentation: one that targets speed without compromising accuracy and the other that targets higher accuracy. The fast version is based on the selection of exemplar image patches from a convolution neural network (CNN) and patch classification by quantifying the divergence between the PHPs of exemplars and the input image patch. Detailed comparative evaluation shows that the proposed algorithm is significantly faster than competing algorithms while achieving comparable results. The accurate version combines the PHPs and high-level CNN features and employs a multi-stage ensemble strategy for image patch labeling. Experimental results demonstrate that the combination of PHPs and CNN features outperforms competing algorithms. This study is performed on two independently collected colorectal datasets containing adenoma, adenocarcinoma, signet and healthy cases. Collectively, the accurate tumor segmentation produces the highest average patch-level F1-score, as compared with competing algorithms, on malignant and healthy cases from both the datasets. Overall the proposed framework highlights the utility of persistent homology for histopathology image analysis.
Micro-Net: A unified model for segmentation of various objects in microscopy images
Apr 22, 2018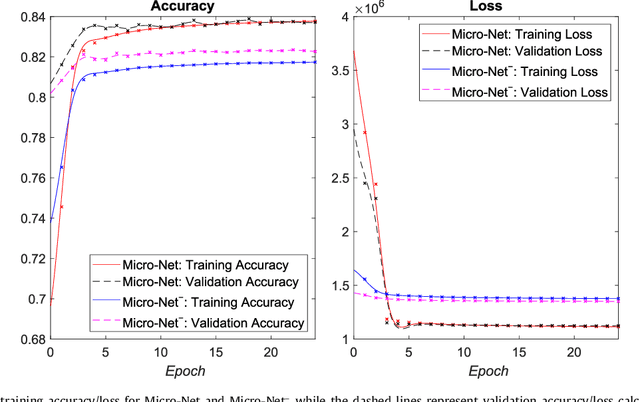
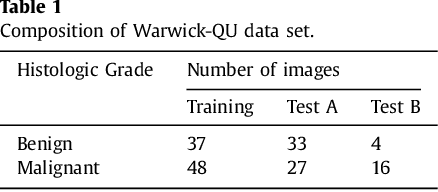
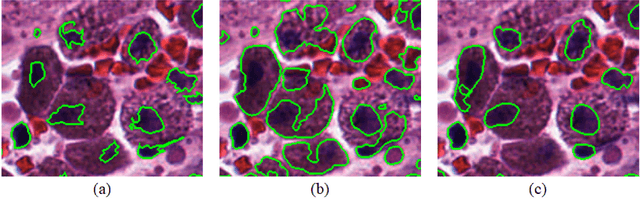
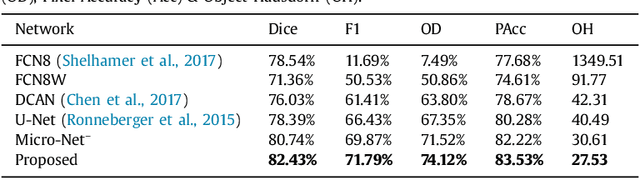
Abstract:Object segmentation and structure localization are important steps in automated image analysis pipelines for microscopy images. We present a convolution neural network (CNN) based deep learning architecture for segmentation of objects in microscopy images. The proposed network can be used to segment cells, nuclei and glands in fluorescence microscopy and histology images after slight tuning of its parameters. It trains itself at multiple resolutions of the input image, connects the intermediate layers for better localization and context and generates the output using multi-resolution deconvolution filters. The extra convolutional layers which bypass the max-pooling operation allow the network to train for variable input intensities and object size and make it robust to noisy data. We compare our results on publicly available data sets and show that the proposed network outperforms the state-of-the-art.
Novel digital tissue phenotypic signatures of distant metastasis in colorectal cancer
Jan 23, 2018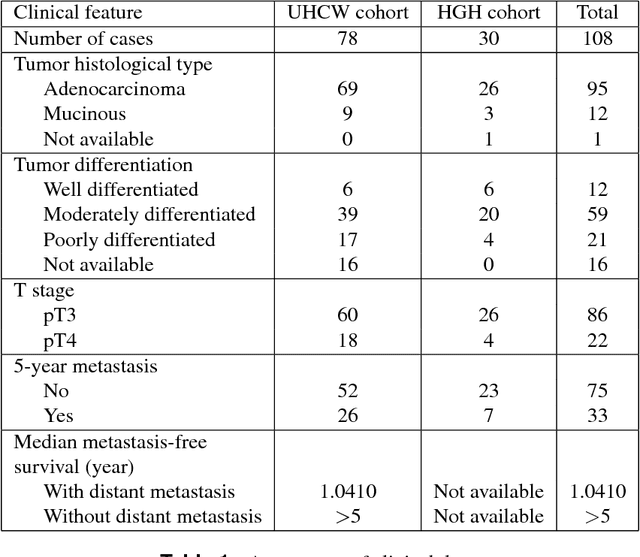
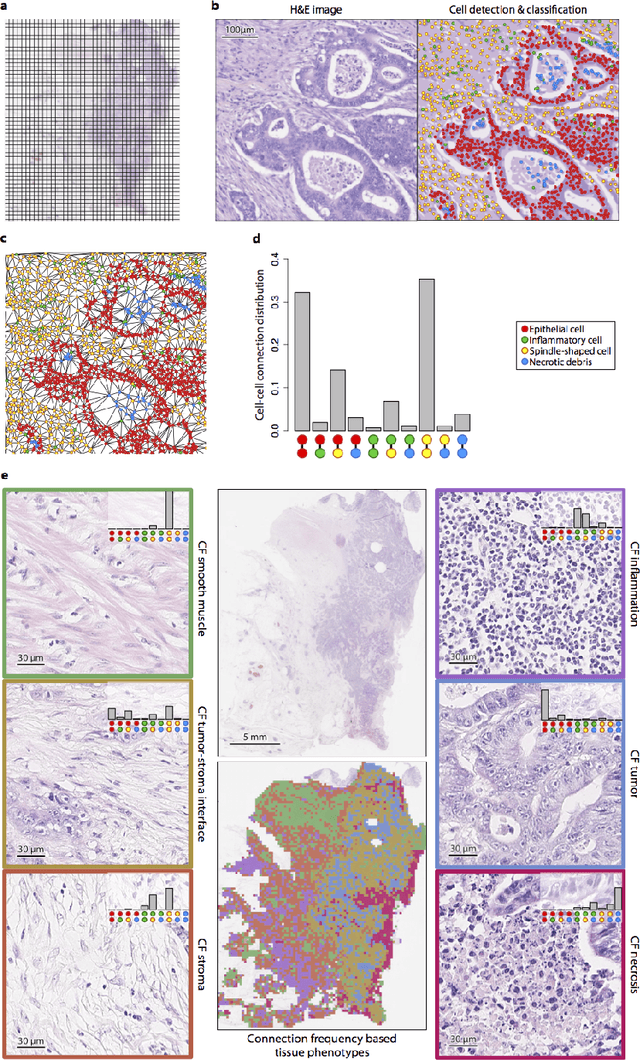
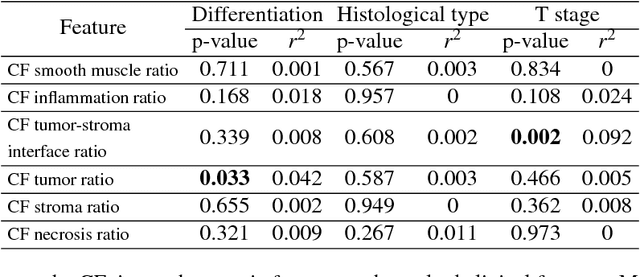
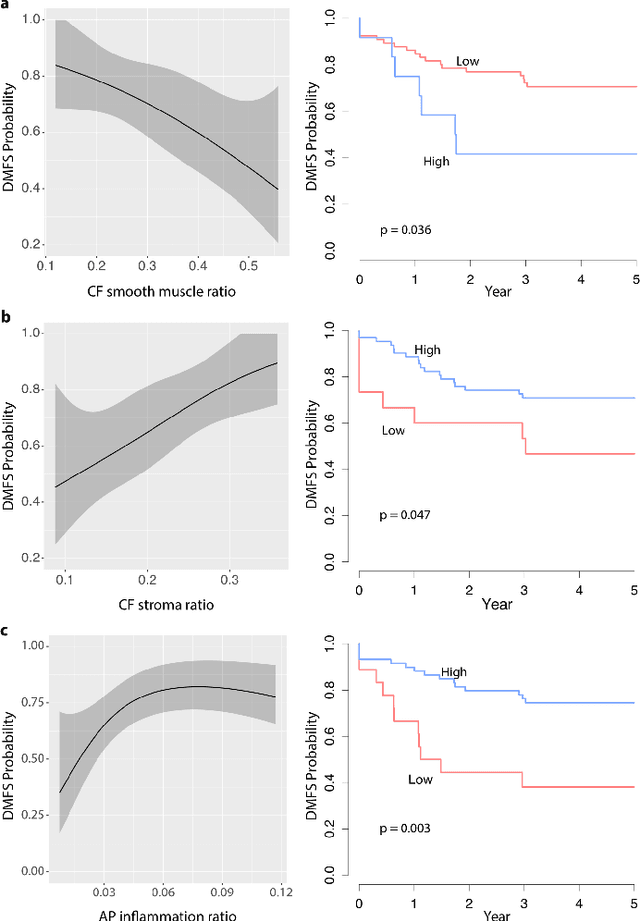
Abstract:Distant metastasis is the major cause of death in colorectal cancer (CRC). Patients at high risk of developing distant metastasis could benefit from appropriate adjuvant and follow-up treatments if stratified accurately at an early stage of the disease. Studies have increasingly recognized the role of diverse cellular components within the tumor microenvironment in the development and progression of CRC tumors. In this paper, we show that a new method of automated analysis of digitized images from colorectal cancer tissue slides can provide important estimates of distant metastasis-free survival (DMFS, the time before metastasis is first observed) on the basis of details of the microenvironment. Specifically, we determine what cell types are found in the vicinity of other cell types, and in what numbers, rather than concentrating exclusively on the cancerous cells. We then extract novel tissue phenotypic signatures using statistical measurements about tissue composition. Such signatures can underpin clinical decisions about the advisability of various types of adjuvant therapy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge