Daniele Perlo
RodEpil: A Video Dataset of Laboratory Rodents for Seizure Detection and Benchmark Evaluation
Nov 14, 2025Abstract:We introduce a curated video dataset of laboratory rodents for automatic detection of convulsive events. The dataset contains short (10~s) top-down and side-view video clips of individual rodents, labeled at clip level as normal activity or seizure. It includes 10,101 negative samples and 2,952 positive samples collected from 19 subjects. We describe the data curation, annotation protocol and preprocessing pipeline, and report baseline experiments using a transformer-based video classifier (TimeSformer). Experiments employ five-fold cross-validation with strict subject-wise partitioning to prevent data leakage (no subject appears in more than one fold). Results show that the TimeSformer architecture enables discrimination between seizure and normal activity with an average F1-score of 97%. The dataset and baseline code are publicly released to support reproducible research on non-invasive, video-based monitoring in preclinical epilepsy research. RodEpil Dataset access - DOI: 10.5281/zenodo.17601357
Exploiting Liver CT scans in Colorectal Carcinoma genomics mutation classification
Jan 25, 2024



Abstract:The liver is the most involved organ by distant metastasis in colon-rectal cancer (CRC) patients and it comes necessary to be aware of the mutational status of the lesions to correctly design the best individual treatment. So far, efforts have been made in order to develop non-invasive and real-time methods that permit the analysis of the whole tumor, using new artificial intelligence tools to analyze the tumor's image obtained by Computed Tomography (CT) scan. In order to address the current medical workflow, that is biopsy analysis-based, we propose the first DeepLearning-based exploration, to our knowledge, of such classification approach from the patient medical imaging. We propose i) a solid pipeline for managing undersized datasets of available CT scans and ii) a baseline study for genomics mutation diagnosis support for preemptive patient follow-up. Our method is able to identify CRC RAS mutation family from CT images with 0.73 F1 score.
Predicting Hypoxia in Brain Tumors from Multiparametric MRI
Jan 25, 2024Abstract:This research paper presents a novel approach to the prediction of hypoxia in brain tumors, using multi-parametric Magnetic Resonance Imaging (MRI). Hypoxia, a condition characterized by low oxygen levels, is a common feature of malignant brain tumors associated with poor prognosis. Fluoromisonidazole Positron Emission Tomography (FMISO PET) is a well-established method for detecting hypoxia in vivo, but it is expensive and not widely available. Our study proposes the use of MRI, a more accessible and cost-effective imaging modality, to predict FMISO PET signals. We investigate deep learning models (DL) trained on the ACRIN 6684 dataset, a resource that contains paired MRI and FMISO PET images from patients with brain tumors. Our trained models effectively learn the complex relationships between the MRI features and the corresponding FMISO PET signals, thereby enabling the prediction of hypoxia from MRI scans alone. The results show a strong correlation between the predicted and actual FMISO PET signals, with an overall PSNR score above 29.6 and a SSIM score greater than 0.94, confirming MRI as a promising option for hypoxia prediction in brain tumors. This approach could significantly improve the accessibility of hypoxia detection in clinical settings, with the potential for more timely and targeted treatments.
UniToBrain dataset: a Brain Perfusion Dataset
Aug 01, 2022Abstract:The CT perfusion (CTP) is a medical exam for measuring the passage of a bolus of contrast solution through the brain on a pixel-by-pixel basis. The objective is to draw "perfusion maps" (namely cerebral blood volume, cerebral blood flow and time to peak) very rapidly for ischemic lesions, and to be able to distinguish between core and penumubra regions. A precise and quick diagnosis, in a context of ischemic stroke, can determine the fate of the brain tissues and guide the intervention and treatment in emergency conditions. In this work we present UniToBrain dataset, the very first open-source dataset for CTP. It comprises a cohort of more than a hundred of patients, and it is accompanied by patients metadata and ground truth maps obtained with state-of-the-art algorithms. We also propose a novel neural networks-based algorithm, using the European library ECVL and EDDL for the image processing and developing deep learning models respectively. The results obtained by the neural network models match the ground truth and open the road towards potential sub-sampling of the required number of CT maps, which impose heavy radiation doses to the patients.
Lung nodules segmentation from CT with DeepHealth toolkit
Aug 01, 2022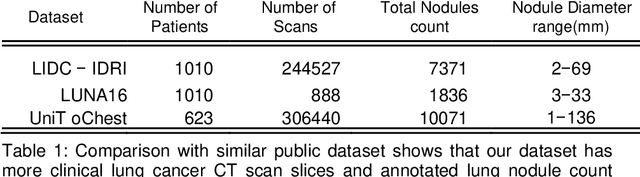
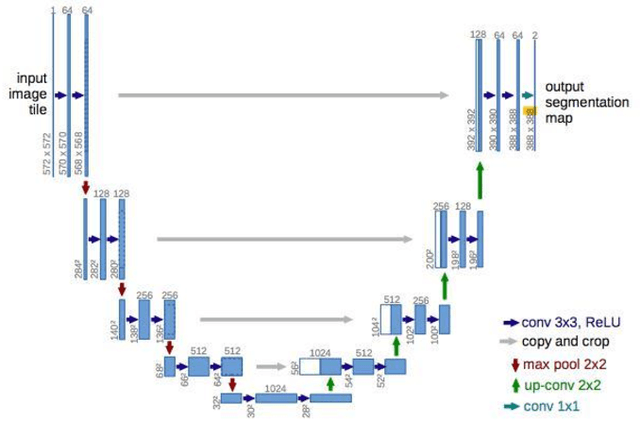

Abstract:The accurate and consistent border segmentation plays an important role in the tumor volume estimation and its treatment in the field of Medical Image Segmentation. Globally, Lung cancer is one of the leading causes of death and the early detection of lung nodules is essential for the early cancer diagnosis and survival rate of patients. The goal of this study was to demonstrate the feasibility of Deephealth toolkit including PyECVL and PyEDDL libraries to precisely segment lung nodules. Experiments for lung nodules segmentation has been carried out on UniToChest using PyECVL and PyEDDL, for data pre-processing as well as neural network training. The results depict accurate segmentation of lung nodules across a wide diameter range and better accuracy over a traditional detection approach. The datasets and the code used in this paper are publicly available as a baseline reference.
Dysplasia grading of colorectal polyps through CNN analysis of WSI
Feb 10, 2021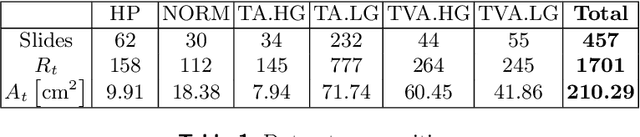
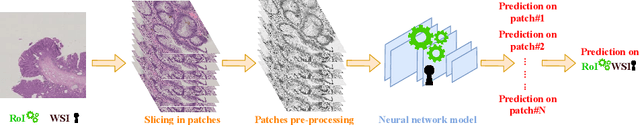
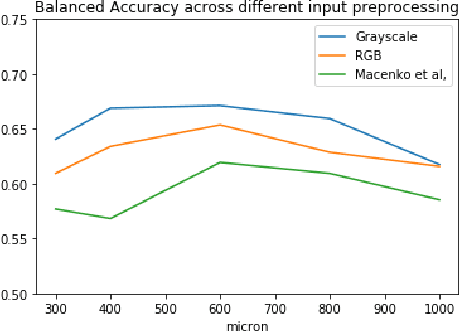
Abstract:Colorectal cancer is a leading cause of cancer death for both men and women. For this reason, histopathological characterization of colorectal polyps is the major instrument for the pathologist in order to infer the actual risk for cancer and to guide further follow-up. Colorectal polyps diagnosis includes the evaluation of the polyp type, and more importantly, the grade of dysplasia. This latter evaluation represents a critical step for the clinical follow-up. The proposed deep learning-based classification pipeline is based on state-of-the-art convolutional neural network, trained using proper countermeasures to tackle WSI high resolution and very imbalanced dataset. The experimental results show that one can successfully classify adenomas dysplasia grade with 70% accuracy, which is in line with the pathologists' concordance.
UniToPatho, a labeled histopathological dataset for colorectal polyps classification and adenoma dysplasia grading
Feb 10, 2021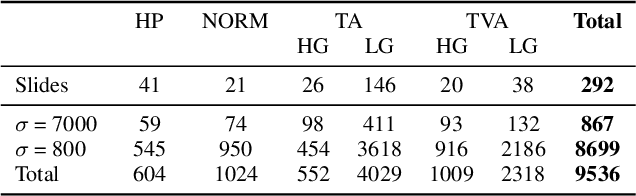
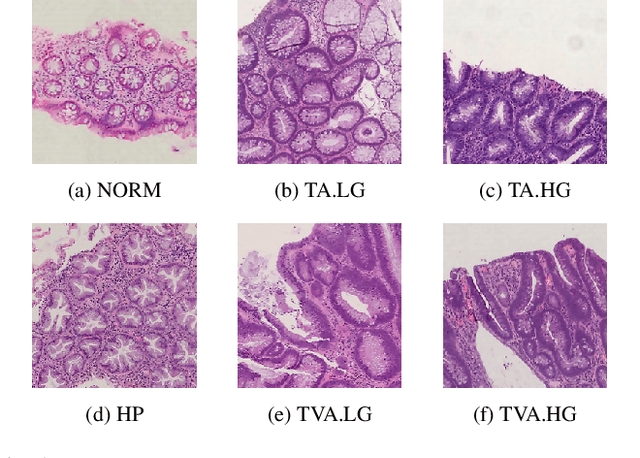
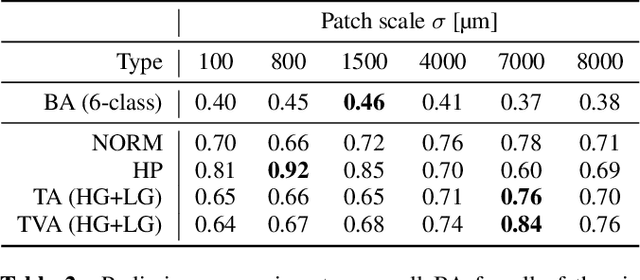
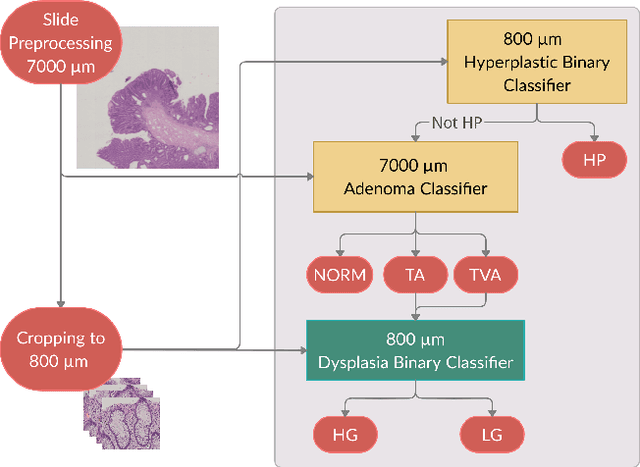
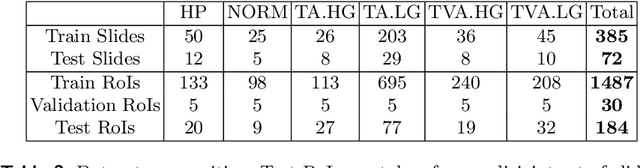
Abstract:Histopathological characterization of colorectal polyps allows to tailor patients' management and follow up with the ultimate aim of avoiding or promptly detecting an invasive carcinoma. Colorectal polyps characterization relies on the histological analysis of tissue samples to determine the polyps malignancy and dysplasia grade. Deep neural networks achieve outstanding accuracy in medical patterns recognition, however they require large sets of annotated training images. We introduce UniToPatho, an annotated dataset of 9536 hematoxylin and eosin (H&E) stained patches extracted from 292 whole-slide images, meant for training deep neural networks for colorectal polyps classification and adenomas grading. We present our dataset and provide insights on how to tackle the problem of automatic colorectal polyps characterization.
Post-synaptic potential regularization has potential
Jul 19, 2019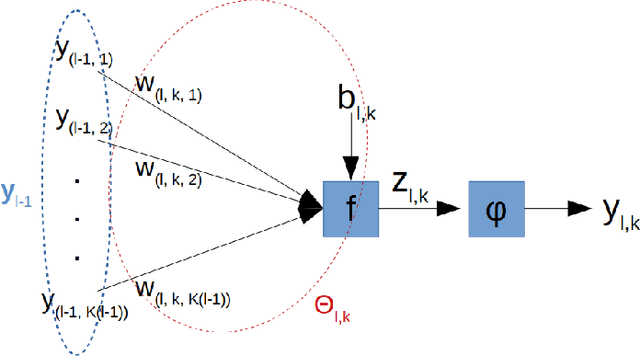

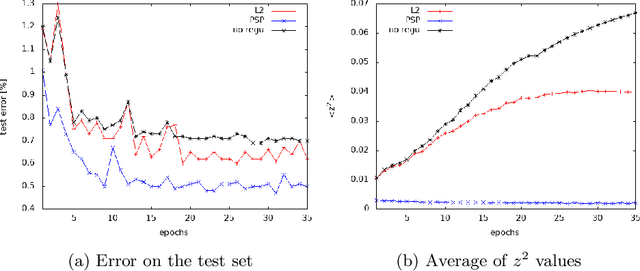

Abstract:Improving generalization is one of the main challenges for training deep neural networks on classification tasks. In particular, a number of techniques have been proposed, aiming to boost the performance on unseen data: from standard data augmentation techniques to the $\ell_2$ regularization, dropout, batch normalization, entropy-driven SGD and many more.\\ In this work we propose an elegant, simple and principled approach: post-synaptic potential regularization (PSP). We tested this regularization on a number of different state-of-the-art scenarios. Empirical results show that PSP achieves a classification error comparable to more sophisticated learning strategies in the MNIST scenario, while improves the generalization compared to $\ell_2$ regularization in deep architectures trained on CIFAR-10.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge