Dani Kiyasseh
Quantification of Robotic Surgeries with Vision-Based Deep Learning
May 06, 2022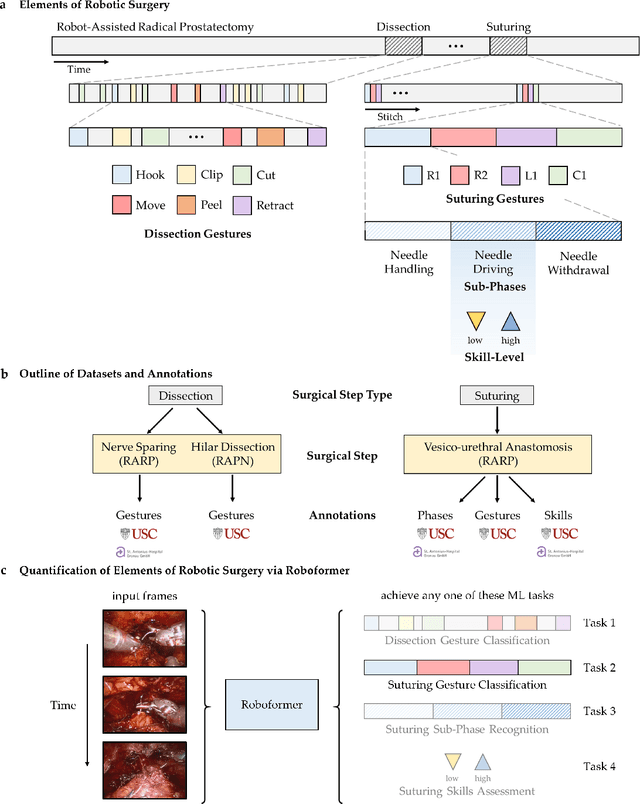
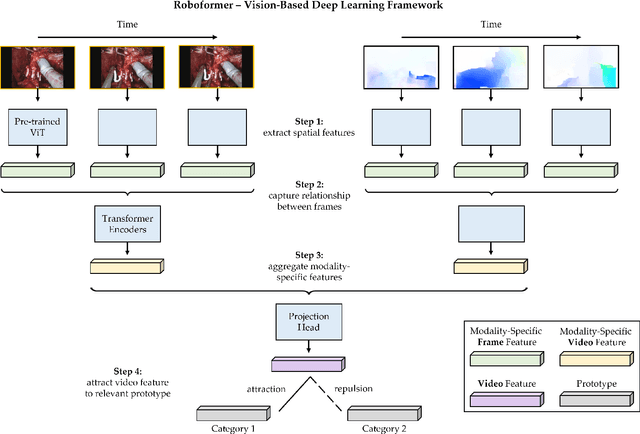
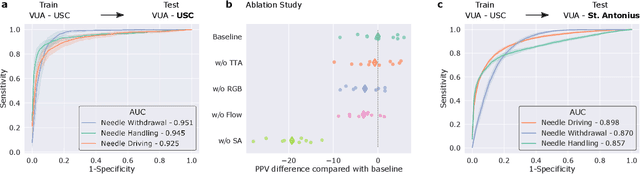
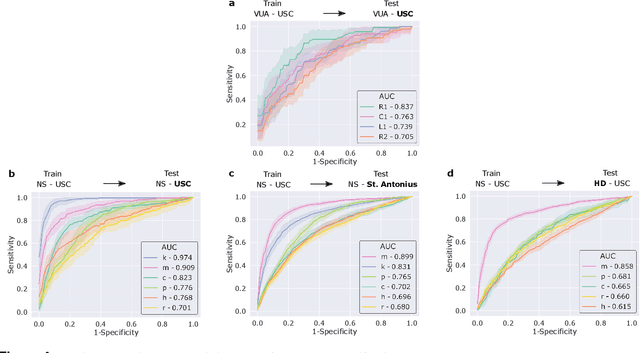
Abstract:Surgery is a high-stakes domain where surgeons must navigate critical anatomical structures and actively avoid potential complications while achieving the main task at hand. Such surgical activity has been shown to affect long-term patient outcomes. To better understand this relationship, whose mechanics remain unknown for the majority of surgical procedures, we hypothesize that the core elements of surgery must first be quantified in a reliable, objective, and scalable manner. We believe this is a prerequisite for the provision of surgical feedback and modulation of surgeon performance in pursuit of improved patient outcomes. To holistically quantify surgeries, we propose a unified deep learning framework, entitled Roboformer, which operates exclusively on videos recorded during surgery to independently achieve multiple tasks: surgical phase recognition (the what of surgery), gesture classification and skills assessment (the how of surgery). We validated our framework on four video-based datasets of two commonly-encountered types of steps (dissection and suturing) within minimally-invasive robotic surgeries. We demonstrated that our framework can generalize well to unseen videos, surgeons, medical centres, and surgical procedures. We also found that our framework, which naturally lends itself to explainable findings, identified relevant information when achieving a particular task. These findings are likely to instill surgeons with more confidence in our framework's behaviour, increasing the likelihood of clinical adoption, and thus paving the way for more targeted surgical feedback.
Turath-150K: Image Database of Arab Heritage
Jan 01, 2022

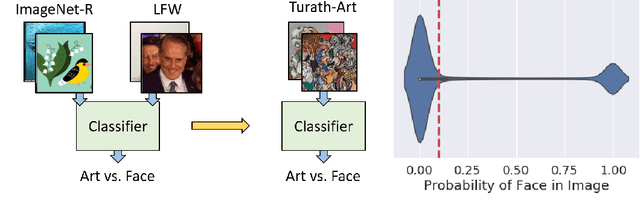

Abstract:Large-scale image databases remain largely biased towards objects and activities encountered in a select few cultures. This absence of culturally-diverse images, which we refer to as the hidden tail, limits the applicability of pre-trained neural networks and inadvertently excludes researchers from under-represented regions. To begin remedying this issue, we curate Turath-150K, a database of images of the Arab world that reflect objects, activities, and scenarios commonly found there. In the process, we introduce three benchmark databases, Turath Standard, Art, and UNESCO, specialised subsets of the Turath dataset. After demonstrating the limitations of existing networks pre-trained on ImageNet when deployed on such benchmarks, we train and evaluate several networks on the task of image classification. As a consequence of Turath, we hope to engage machine learning researchers in under-represented regions, and to inspire the release of additional culture-focused databases. The database can be accessed here: danikiyasseh.github.io/Turath.
Let Your Heart Speak in its Mother Tongue: Multilingual Captioning of Cardiac Signals
Mar 19, 2021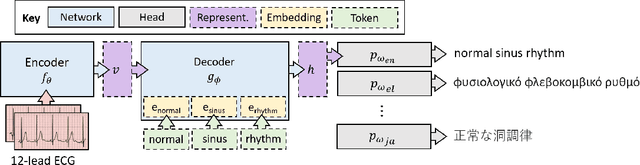

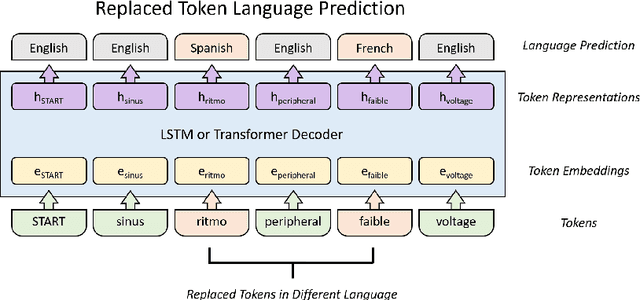

Abstract:Cardiac signals, such as the electrocardiogram, convey a significant amount of information about the health status of a patient which is typically summarized by a clinician in the form of a clinical report, a cumbersome process that is prone to errors. To streamline this routine process, we propose a deep neural network capable of captioning cardiac signals; it receives a cardiac signal as input and generates a clinical report as output. We extend this further to generate multilingual reports. To that end, we create and make publicly available a multilingual clinical report dataset. In the absence of sufficient labelled data, deep neural networks can benefit from a warm-start, or pre-training, procedure in which parameters are first learned in an arbitrary task. We propose such a task in the form of discriminative multilingual pre-training where tokens from clinical reports are randomly replaced with those from other languages and the network is tasked with predicting the language of all tokens. We show that our method performs on par with state-of-the-art pre-training methods such as MLM, ELECTRA, and MARGE, while simultaneously generating diverse and plausible clinical reports. We also demonstrate that multilingual models can outperform their monolingual counterparts, informally terming this beneficial phenomenon as the blessing of multilinguality.
DROPS: Deep Retrieval of Physiological Signals via Attribute-specific Clinical Prototypes
Nov 28, 2020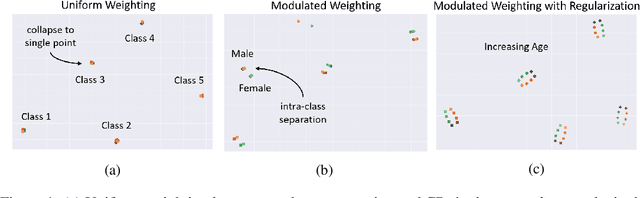
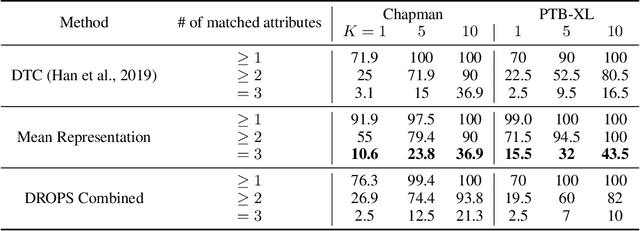
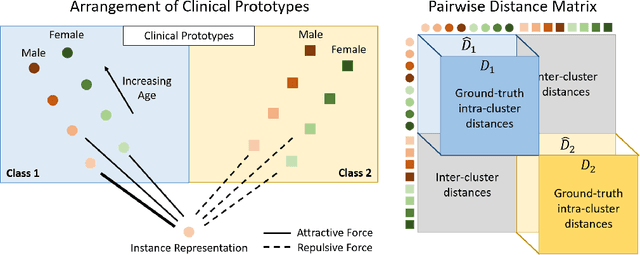
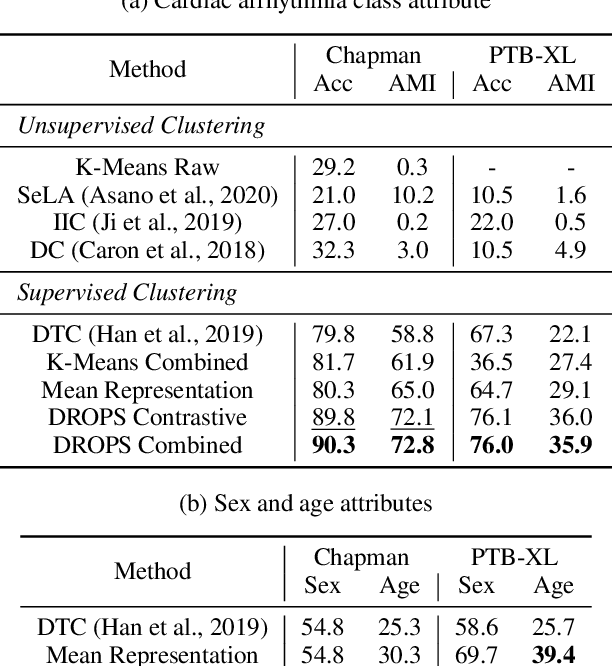
Abstract:The ongoing digitization of health records within the healthcare industry results in large-scale datasets. Manually extracting clinically-useful insight from such datasets is non-trivial. However, doing so at scale while simultaneously leveraging patient-specific attributes such as sex and age can assist with clinical-trial enrollment, medical school educational endeavours, and the evaluation of the fairness of neural networks. To facilitate the reliable extraction of clinical information, we propose to learn embeddings, known as clinical prototypes (CPs), via supervised contrastive learning. We show that CPs can be efficiently used for large-scale retrieval and clustering of physiological signals based on multiple patient attributes. We also show that CPs capture attribute-specific semantic relationships.
PCPs: Patient Cardiac Prototypes
Nov 28, 2020
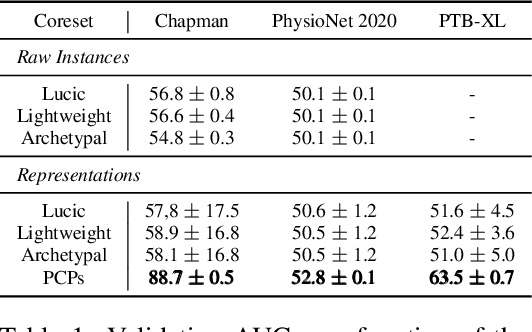
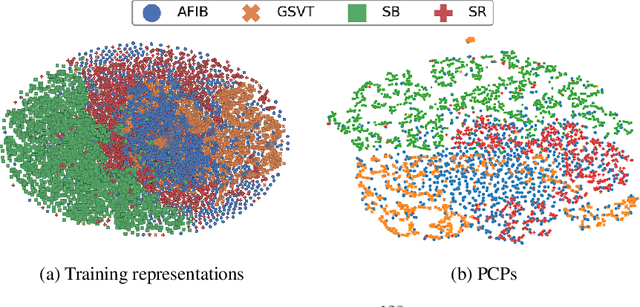
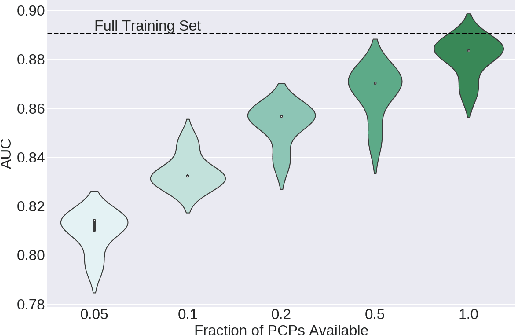
Abstract:Many clinical deep learning algorithms are population-based and difficult to interpret. Such properties limit their clinical utility as population-based findings may not generalize to individual patients and physicians are reluctant to incorporate opaque models into their clinical workflow. To overcome these obstacles, we propose to learn patient-specific embeddings, entitled patient cardiac prototypes (PCPs), that efficiently summarize the cardiac state of the patient. To do so, we attract representations of multiple cardiac signals from the same patient to the corresponding PCP via supervised contrastive learning. We show that the utility of PCPs is multifold. First, they allow for the discovery of similar patients both within and across datasets. Second, such similarity can be leveraged in conjunction with a hypernetwork to generate patient-specific parameters, and in turn, patient-specific diagnoses. Third, we find that PCPs act as a compact substitute for the original dataset, allowing for dataset distillation.
CLOCS: Contrastive Learning of Cardiac Signals
May 27, 2020
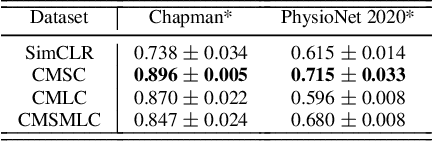
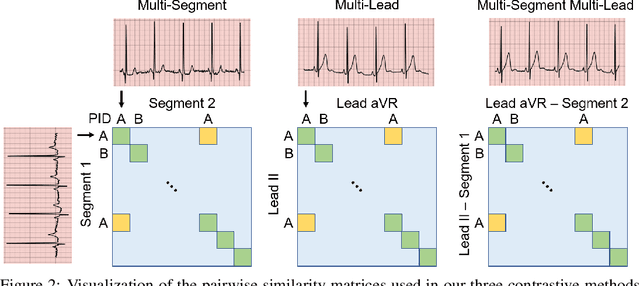
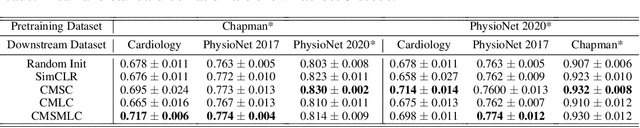
Abstract:The healthcare industry generates troves of unlabelled physiological data. This data can be exploited via contrastive learning, a self-supervised pre-training mechanism that encourages representations of instances to be similar to one another. We propose a family of contrastive learning methods, CLOCS, that encourages representations across time, leads, and patients to be similar to one another. We show that CLOCS consistently outperforms the state-of-the-art approach, SimCLR, on both linear evaluation and fine-tuning downstream tasks. We also show that CLOCS achieves strong generalization performance with only 25% of labelled training data. Furthermore, our training procedure naturally generates patient-specific representations that can be used to quantify patient-similarity.
SoQal: Selective Oracle Questioning in Active Learning
Apr 22, 2020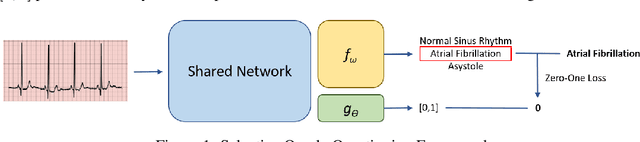
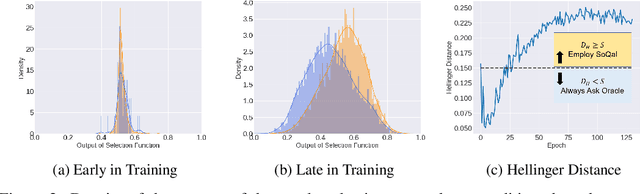

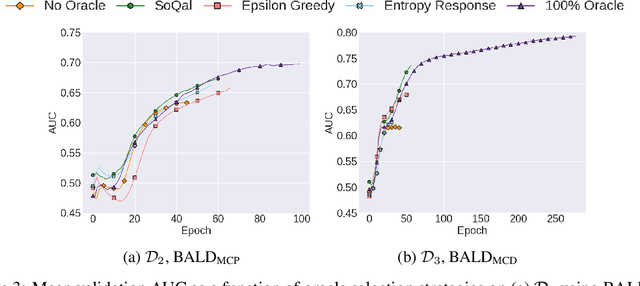
Abstract:Large sets of unlabelled data within the healthcare domain remain underutilized. Active learning offers a way to exploit these datasets by iteratively requesting an oracle (e.g. medical professional) to label instances. This process, which can be costly and time-consuming is overly-dependent upon an oracle. To alleviate this burden, we propose SoQal, a questioning strategy that dynamically determines when a label should be requested from an oracle. We perform experiments on five publically-available datasets and illustrate SoQal's superiority relative to baseline approaches, including its ability to reduce oracle label requests by up to 35%. SoQal also performs competitively in the presence of label noise: a scenario that simulates clinicians' uncertain diagnoses when faced with difficult classification tasks.
CLOPS: Continual Learning of Physiological Signals
Apr 20, 2020
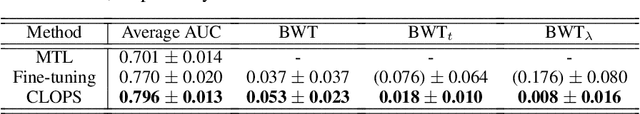
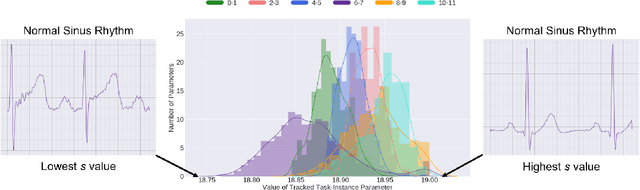

Abstract:Deep learning algorithms are known to experience destructive interference when instances violate the assumption of being independent and identically distributed (i.i.d). This violation, however, is ubiquitous in clinical settings where data are streamed temporally and from a multitude of physiological sensors. To overcome this obstacle, we propose CLOPS, a healthcare-specific replay-based continual learning strategy. In three continual learning scenarios based on three publically-available datasets, we show that CLOPS can outperform its multi-task learning counterpart. Moreover, we propose end-to-end trainable parameters, which we term task-instance parameters, that can be used to quantify task difficulty and similarity. This quantification yields insights into both network interpretability and clinical applications, where task difficulty is poorly quantified.
ALPS: Active Learning via Perturbations
Apr 20, 2020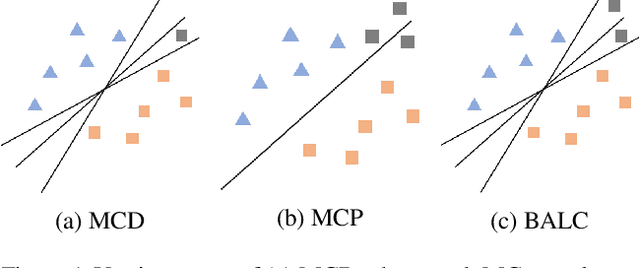
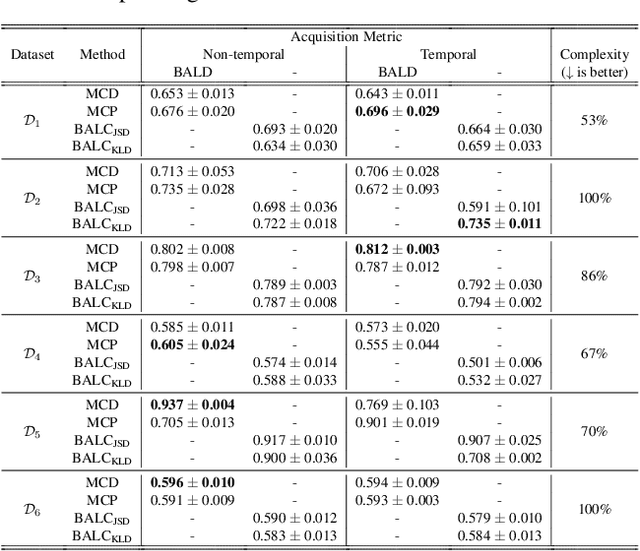
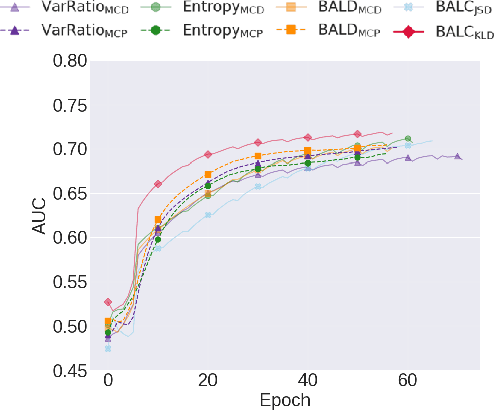
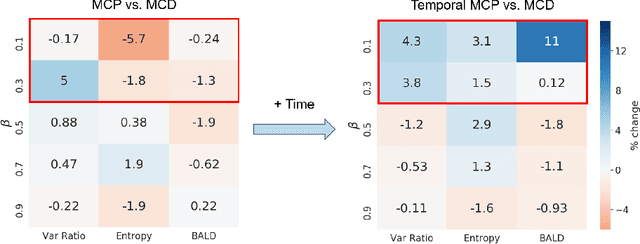
Abstract:Small, labelled datasets in the presence of larger, unlabelled datasets pose challenges to data-hungry deep learning algorithms. Such scenarios are prevalent in healthcare where labelling is expensive, time-consuming, and requires expert medical professionals. To tackle this challenge, we propose a family of active learning methodologies and acquisition functions dependent upon input and parameter perturbations which we call Active Learning via Perturbations (ALPS). We test our methods on six diverse time-series and image datasets and illustrate their benefit in the presence and absence of an oracle. We also show that acquisition functions that incorporate temporal information have the potential to predict the ability of networks to generalize.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge