Dan Adam
High frame-rate cardiac ultrasound imaging with deep learning
Aug 23, 2018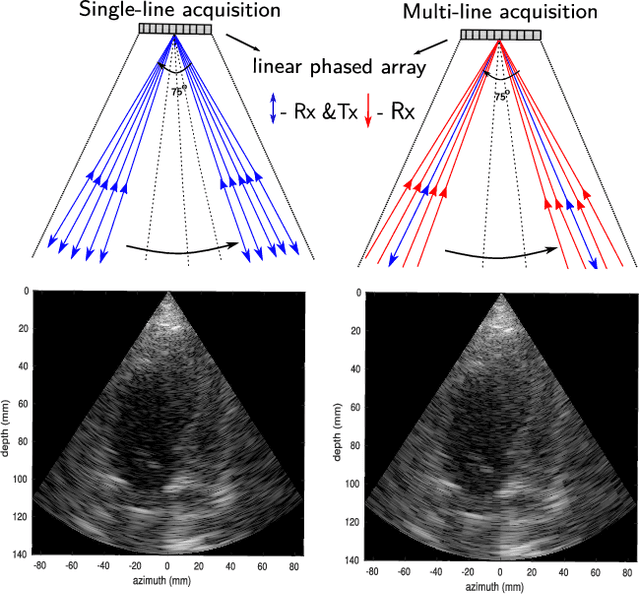

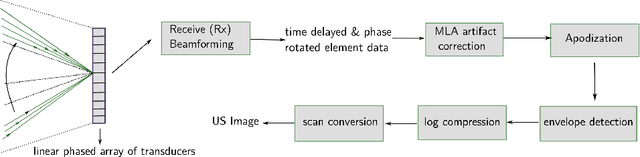

Abstract:Cardiac ultrasound imaging requires a high frame rate in order to capture rapid motion. This can be achieved by multi-line acquisition (MLA), where several narrow-focused received lines are obtained from each wide-focused transmitted line. This shortens the acquisition time at the expense of introducing block artifacts. In this paper, we propose a data-driven learning-based approach to improve the MLA image quality. We train an end-to-end convolutional neural network on pairs of real ultrasound cardiac data, acquired through MLA and the corresponding single-line acquisition (SLA). The network achieves a significant improvement in image quality for both $5-$ and $7-$line MLA resulting in a decorrelation measure similar to that of SLA while having the frame rate of MLA.
High quality ultrasonic multi-line transmission through deep learning
Aug 23, 2018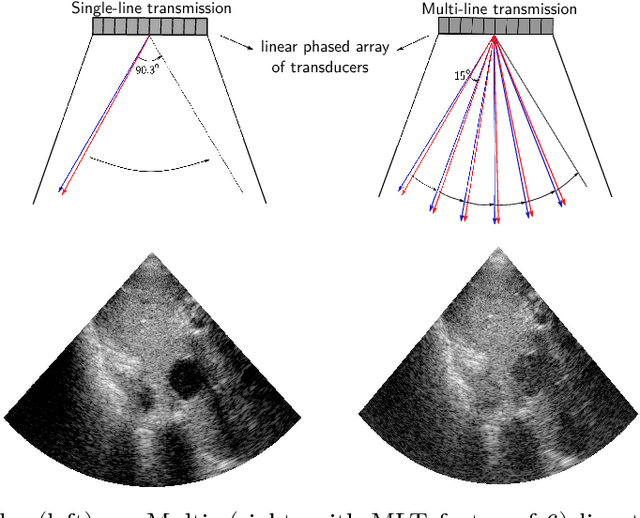
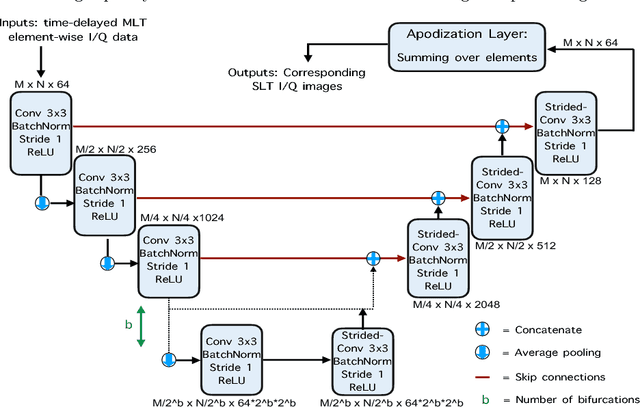
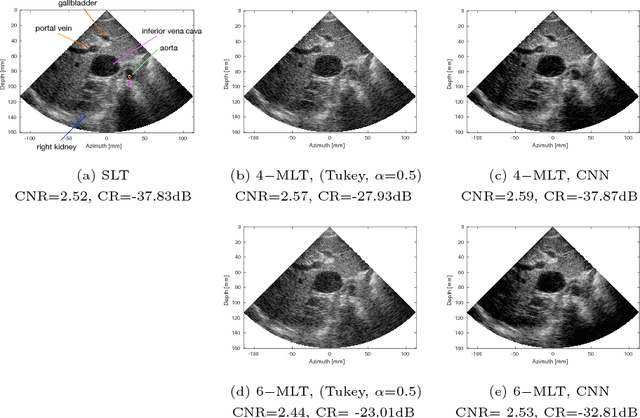
Abstract:Frame rate is a crucial consideration in cardiac ultrasound imaging and 3D sonography. Several methods have been proposed in the medical ultrasound literature aiming at accelerating the image acquisition. In this paper, we consider one such method called \textit{multi-line transmission} (MLT), in which several evenly separated focused beams are transmitted simultaneously. While MLT reduces the acquisition time, it comes at the expense of a heavy loss of contrast due to the interactions between the beams (cross-talk artifact). In this paper, we introduce a data-driven method to reduce the artifacts arising in MLT. To this end, we propose to train an end-to-end convolutional neural network consisting of correction layers followed by a constant apodization layer. The network is trained on pairs of raw data obtained through MLT and the corresponding \textit{single-line transmission} (SLT) data. Experimental evaluation demonstrates significant improvement both in the visual image quality and in objective measures such as contrast ratio and contrast-to-noise ratio, while preserving resolution unlike traditional apodization-based methods. We show that the proposed method is able to generalize well across different patients and anatomies on real and phantom data.
Using Anatomical Markers for Left Ventricular Segmentation of Long Axis Ultrasound Images
Oct 12, 2015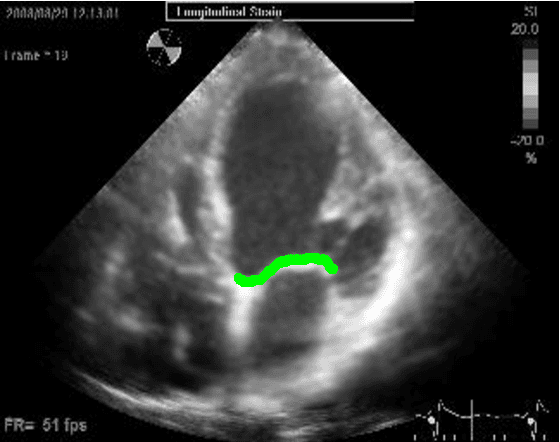
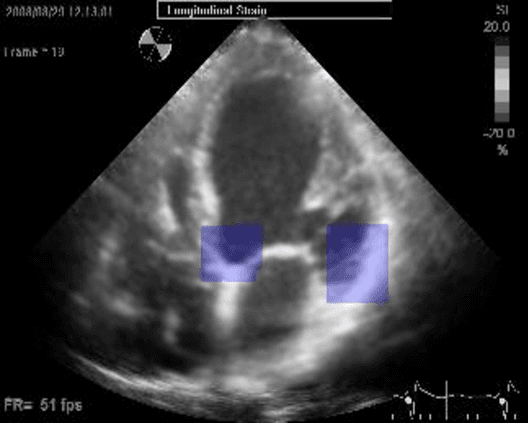
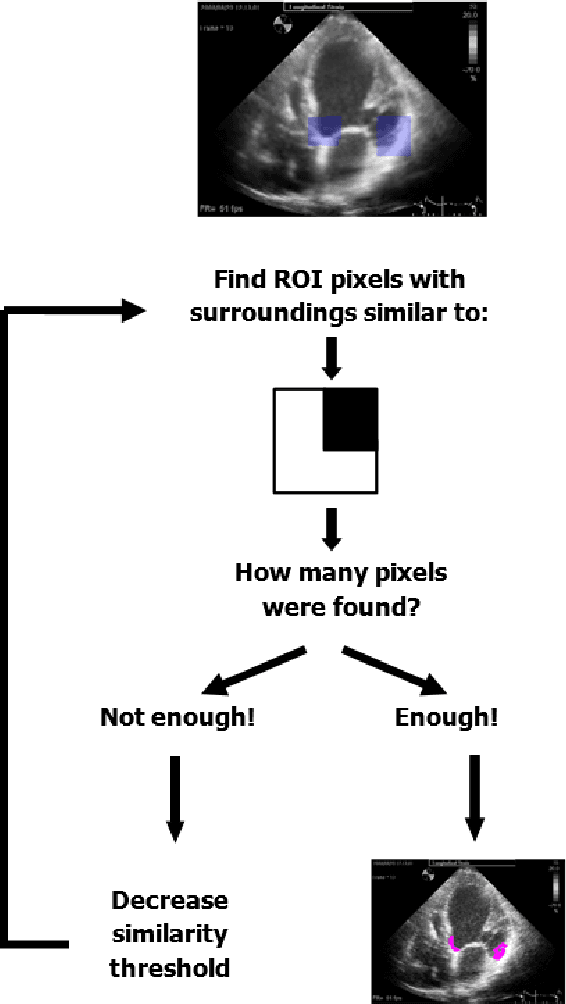
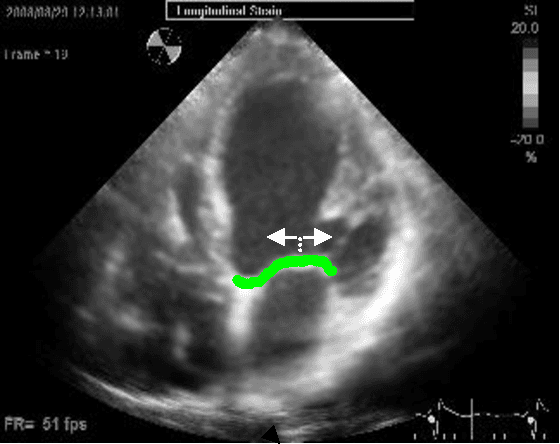
Abstract:Left ventricular segmentation is essential for measuring left ventricular function indices. Segmentation of one or several images requires an initial guess of the contour. It is hypothesized here that creating an initial guess by first detecting anatomical markers, would lead to correct detection of the endocardium. The first step of the algorithm presented here includes automatic detection of the mitral valve. Next, the apex is detected in the same frame. The valve is then tracked throughout the cardiac cycle. Contours passing from the apex to each valve corner are then found using a dynamic programming algorithm. The resulting contour is used as an input to an active contour algorithm. The algorithm was tested on 21 long axis ultrasound clips and showed good agreement with manually traced contours. Thus, this study demonstrates that detection of anatomic markers leads to a reliable initial guess of the left ventricle border.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge