Christophe Guéret
Veni, Vidi, Vici: Solving the Myriad of Challenges before Knowledge Graph Learning
Feb 08, 2024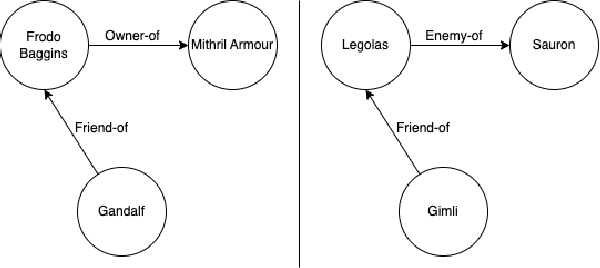
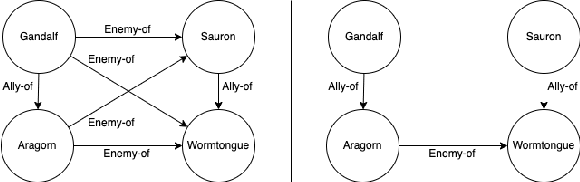
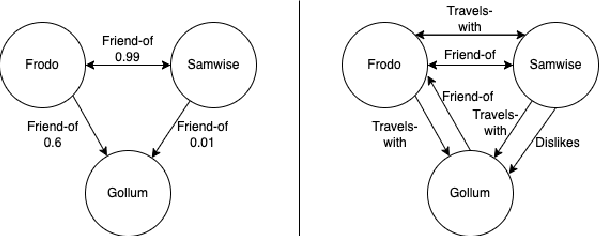
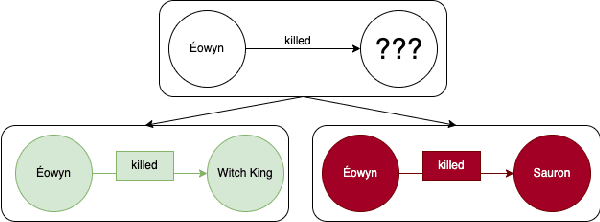
Abstract:Knowledge Graphs (KGs) have become increasingly common for representing large-scale linked data. However, their immense size has required graph learning systems to assist humans in analysis, interpretation, and pattern detection. While there have been promising results for researcher- and clinician- empowerment through a variety of KG learning systems, we identify four key deficiencies in state-of-the-art graph learning that simultaneously limit KG learning performance and diminish the ability of humans to interface optimally with these learning systems. These deficiencies are: 1) lack of expert knowledge integration, 2) instability to node degree extremity in the KG, 3) lack of consideration for uncertainty and relevance while learning, and 4) lack of explainability. Furthermore, we characterise state-of-the-art attempts to solve each of these problems and note that each attempt has largely been isolated from attempts to solve the other problems. Through a formalisation of these problems and a review of the literature that addresses them, we adopt the position that not only are deficiencies in these four key areas holding back human-KG empowerment, but that the divide-and-conquer approach to solving these problems as individual units rather than a whole is a significant barrier to the interface between humans and KG learning systems. We propose that it is only through integrated, holistic solutions to the limitations of KG learning systems that human and KG learning co-empowerment will be efficiently affected. We finally present our "Veni, Vidi, Vici" framework that sets a roadmap for effectively and efficiently shifting to a holistic co-empowerment model in both the KG learning and the broader machine learning domain.
Predicting Illness for a Sustainable Dairy Agriculture: Predicting and Explaining the Onset of Mastitis in Dairy Cows
Jan 07, 2021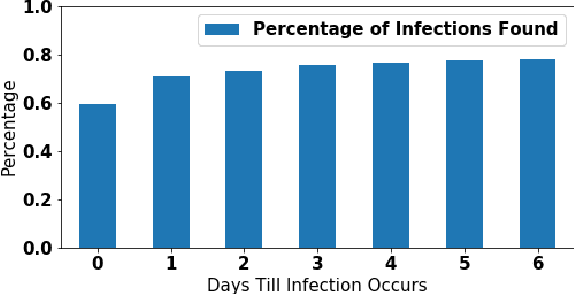
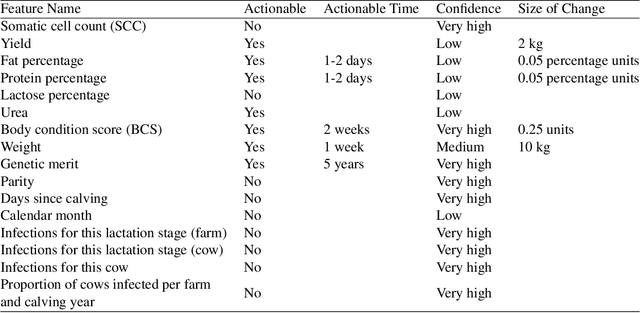
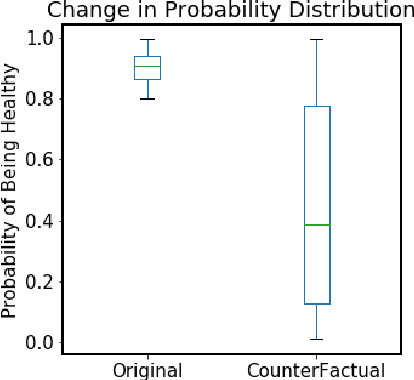

Abstract:Mastitis is a billion dollar health problem for the modern dairy industry, with implications for antibiotic resistance. The use of AI techniques to identify the early onset of this disease, thus has significant implications for the sustainability of this agricultural sector. Current approaches to treating mastitis involve antibiotics and this practice is coming under ever increasing scrutiny. Using machine learning models to identify cows at risk of developing mastitis and applying targeted treatment regimes to only those animals promotes a more sustainable approach. Incorrect predictions from such models, however, can lead to monetary losses, unnecessary use of antibiotics, and even the premature death of animals, so it is important to generate compelling explanations for predictions to build trust with users and to better support their decision making. In this paper we demonstrate a system developed to predict mastitis infections in cows and provide explanations of these predictions using counterfactuals. We demonstrate the system and describe the engagement with farmers undertaken to build it.
Can We Detect Mastitis earlier than Farmers?
Nov 05, 2020


Abstract:The aim of this study was to build a modelling framework that would allow us to be able to detect mastitis infections before they would normally be found by farmers through the introduction of machine learning techniques. In the making of this we created two different modelling framework's, one that works on the premise of detecting Sub Clinical mastitis infections at one Somatic Cell Count recording in advance called SMA and the other tries to detect both Sub Clinical mastitis infections aswell as Clinical mastitis infections at any time the cow is milked called AMA. We also introduce the idea of two different feature sets for our study, these represent different characteristics that should be taken into account when detecting infections, these were the idea of a cow differing to a farm mean and also trends in the lactation. We reported that the results for SMA are better than those created by AMA for Sub Clinical infections yet it has the significant disadvantage of only being able to classify Sub Clinical infections due to how we recorded Sub Clinical infections as being any time a Somatic Cell Count measurement went above a certain threshold where as CM could appear at any stage of lactation. Thus in some cases the lower accuracy values for AMA might in fact be more beneficial to farmers.
Release Early, Release Often: Predicting Change in Versioned Knowledge Organization Systems on the Web
Sep 15, 2015



Abstract:The Semantic Web is built on top of Knowledge Organization Systems (KOS) (vocabularies, ontologies, concept schemes) that provide a structured, interoperable and distributed access to Linked Data on the Web. The maintenance of these KOS over time has produced a number of KOS version chains: subsequent unique version identifiers to unique states of a KOS. However, the release of new KOS versions pose challenges to both KOS publishers and users. For publishers, updating a KOS is a knowledge intensive task that requires a lot of manual effort, often implying deep deliberation on the set of changes to introduce. For users that link their datasets to these KOS, a new version compromises the validity of their links, often creating ramifications. In this paper we describe a method to automatically detect which parts of a Web KOS are likely to change in a next version, using supervised learning on past versions in the KOS version chain. We use a set of ontology change features to model and predict change in arbitrary Web KOS. We apply our method on 139 varied datasets systematically retrieved from the Semantic Web, obtaining robust results at correctly predicting change. To illustrate the accuracy, genericity and domain independence of the method, we study the relationship between its effectiveness and several characterizations of the evaluated datasets, finding that predictors like the number of versions in a chain and their release frequency have a fundamental impact in predictability of change in Web KOS. Consequently, we argue for adopting a release early, release often philosophy in Web KOS development cycles.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge