Chixiang Lu
GraVIS: Grouping Augmented Views from Independent Sources for Dermatology Analysis
Jan 11, 2023
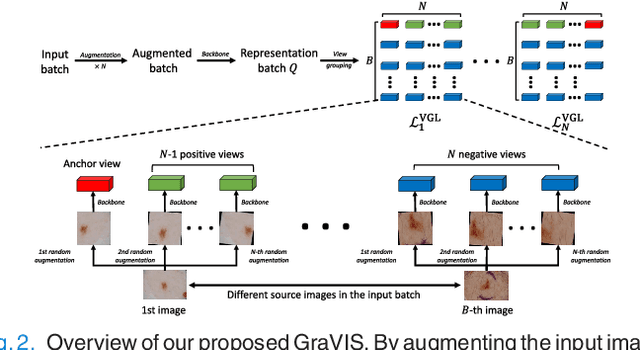


Abstract:Self-supervised representation learning has been extremely successful in medical image analysis, as it requires no human annotations to provide transferable representations for downstream tasks. Recent self-supervised learning methods are dominated by noise-contrastive estimation (NCE, also known as contrastive learning), which aims to learn invariant visual representations by contrasting one homogeneous image pair with a large number of heterogeneous image pairs in each training step. Nonetheless, NCE-based approaches still suffer from one major problem that is one homogeneous pair is not enough to extract robust and invariant semantic information. Inspired by the archetypical triplet loss, we propose GraVIS, which is specifically optimized for learning self-supervised features from dermatology images, to group homogeneous dermatology images while separating heterogeneous ones. In addition, a hardness-aware attention is introduced and incorporated to address the importance of homogeneous image views with similar appearance instead of those dissimilar homogeneous ones. GraVIS significantly outperforms its transfer learning and self-supervised learning counterparts in both lesion segmentation and disease classification tasks, sometimes by 5 percents under extremely limited supervision. More importantly, when equipped with the pre-trained weights provided by GraVIS, a single model could achieve better results than winners that heavily rely on ensemble strategies in the well-known ISIC 2017 challenge.
PCRLv2: A Unified Visual Information Preservation Framework for Self-supervised Pre-training in Medical Image Analysis
Jan 02, 2023Abstract:Recent advances in self-supervised learning (SSL) in computer vision are primarily comparative, whose goal is to preserve invariant and discriminative semantics in latent representations by comparing siamese image views. However, the preserved high-level semantics do not contain enough local information, which is vital in medical image analysis (e.g., image-based diagnosis and tumor segmentation). To mitigate the locality problem of comparative SSL, we propose to incorporate the task of pixel restoration for explicitly encoding more pixel-level information into high-level semantics. We also address the preservation of scale information, a powerful tool in aiding image understanding but has not drawn much attention in SSL. The resulting framework can be formulated as a multi-task optimization problem on the feature pyramid. Specifically, we conduct multi-scale pixel restoration and siamese feature comparison in the pyramid. In addition, we propose non-skip U-Net to build the feature pyramid and develop sub-crop to replace multi-crop in 3D medical imaging. The proposed unified SSL framework (PCRLv2) surpasses its self-supervised counterparts on various tasks, including brain tumor segmentation (BraTS 2018), chest pathology identification (ChestX-ray, CheXpert), pulmonary nodule detection (LUNA), and abdominal organ segmentation (LiTS), sometimes outperforming them by large margins with limited annotations.
Preservational Learning Improves Self-supervised Medical Image Models by Reconstructing Diverse Contexts
Sep 09, 2021



Abstract:Preserving maximal information is one of principles of designing self-supervised learning methodologies. To reach this goal, contrastive learning adopts an implicit way which is contrasting image pairs. However, we believe it is not fully optimal to simply use the contrastive estimation for preservation. Moreover, it is necessary and complemental to introduce an explicit solution to preserve more information. From this perspective, we introduce Preservational Learning to reconstruct diverse image contexts in order to preserve more information in learned representations. Together with the contrastive loss, we present Preservational Contrastive Representation Learning (PCRL) for learning self-supervised medical representations. PCRL provides very competitive results under the pretraining-finetuning protocol, outperforming both self-supervised and supervised counterparts in 5 classification/segmentation tasks substantially.
ConvNets vs. Transformers: Whose Visual Representations are More Transferable?
Aug 17, 2021



Abstract:Vision transformers have attracted much attention from computer vision researchers as they are not restricted to the spatial inductive bias of ConvNets. However, although Transformer-based backbones have achieved much progress on ImageNet classification, it is still unclear whether the learned representations are as transferable as or even more transferable than ConvNets' features. To address this point, we systematically investigate the transfer learning ability of ConvNets and vision transformers in 15 single-task and multi-task performance evaluations. Given the strong correlation between the performance of pre-trained models and transfer learning, we include 2 residual ConvNets (i.e., R-101x3 and R-152x4) and 3 Transformer-based visual backbones (i.e., ViT-B, ViT-L and Swin-B), which have close error rates on ImageNet, that indicate similar transfer learning performance on downstream datasets. We observe consistent advantages of Transformer-based backbones on 13 downstream tasks (out of 15), including but not limited to fine-grained classification, scene recognition (classification, segmentation and depth estimation), open-domain classification, face recognition, etc. More specifically, we find that two ViT models heavily rely on whole network fine-tuning to achieve performance gains while Swin Transformer does not have such a requirement. Moreover, vision transformers behave more robustly in multi-task learning, i.e., bringing more improvements when managing mutually beneficial tasks and reducing performance losses when tackling irrelevant tasks. We hope our discoveries can facilitate the exploration and exploitation of vision transformers in the future.
Generalized Organ Segmentation by Imitating One-shot Reasoning using Anatomical Correlation
Mar 30, 2021



Abstract:Learning by imitation is one of the most significant abilities of human beings and plays a vital role in human's computational neural system. In medical image analysis, given several exemplars (anchors), experienced radiologist has the ability to delineate unfamiliar organs by imitating the reasoning process learned from existing types of organs. Inspired by this observation, we propose OrganNet which learns a generalized organ concept from a set of annotated organ classes and then transfer this concept to unseen classes. In this paper, we show that such process can be integrated into the one-shot segmentation task which is a very challenging but meaningful topic. We propose pyramid reasoning modules (PRMs) to model the anatomical correlation between anchor and target volumes. In practice, the proposed module first computes a correlation matrix between target and anchor computerized tomography (CT) volumes. Then, this matrix is used to transform the feature representations of both anchor volume and its segmentation mask. Finally, OrganNet learns to fuse the representations from various inputs and predicts segmentation results for target volume. Extensive experiments show that OrganNet can effectively resist the wide variations in organ morphology and produce state-of-the-art results in one-shot segmentation task. Moreover, even when compared with fully-supervised segmentation models, OrganNet is still able to produce satisfying segmentation results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge