Cheekong Lee
ProtGNN: Towards Self-Explaining Graph Neural Networks
Dec 02, 2021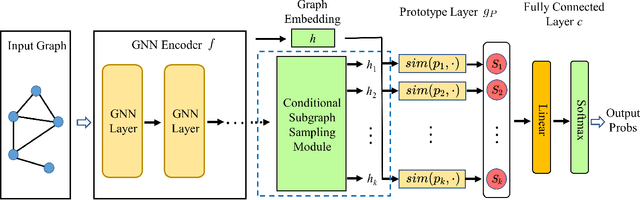
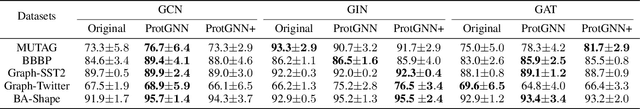
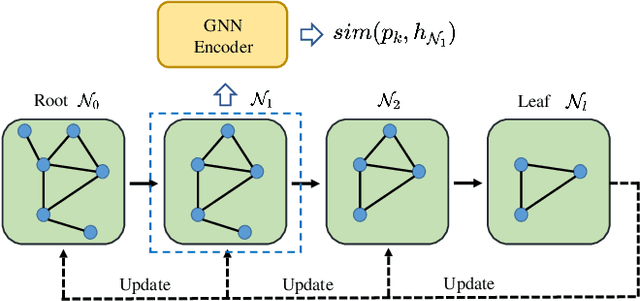

Abstract:Despite the recent progress in Graph Neural Networks (GNNs), it remains challenging to explain the predictions made by GNNs. Existing explanation methods mainly focus on post-hoc explanations where another explanatory model is employed to provide explanations for a trained GNN. The fact that post-hoc methods fail to reveal the original reasoning process of GNNs raises the need of building GNNs with built-in interpretability. In this work, we propose Prototype Graph Neural Network (ProtGNN), which combines prototype learning with GNNs and provides a new perspective on the explanations of GNNs. In ProtGNN, the explanations are naturally derived from the case-based reasoning process and are actually used during classification. The prediction of ProtGNN is obtained by comparing the inputs to a few learned prototypes in the latent space. Furthermore, for better interpretability and higher efficiency, a novel conditional subgraph sampling module is incorporated to indicate which part of the input graph is most similar to each prototype in ProtGNN+. Finally, we evaluate our method on a wide range of datasets and perform concrete case studies. Extensive results show that ProtGNN and ProtGNN+ can provide inherent interpretability while achieving accuracy on par with the non-interpretable counterparts.
ASGN: An Active Semi-supervised Graph Neural Network for Molecular Property Prediction
Jul 07, 2020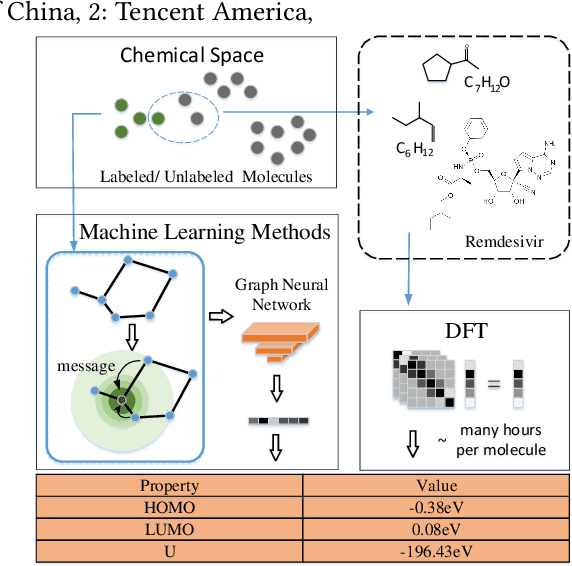
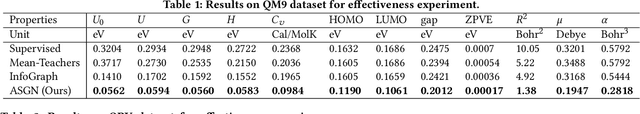
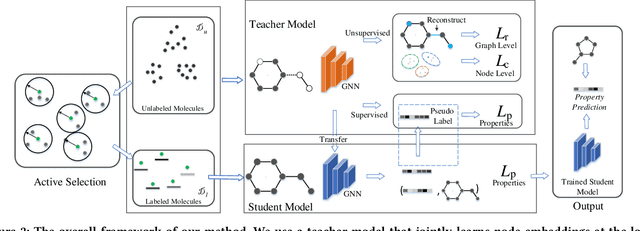
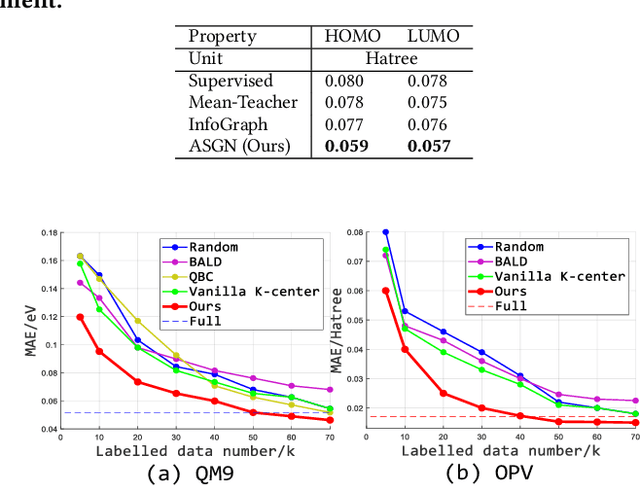
Abstract:Molecular property prediction (e.g., energy) is an essential problem in chemistry and biology. Unfortunately, many supervised learning methods usually suffer from the problem of scarce labeled molecules in the chemical space, where such property labels are generally obtained by Density Functional Theory (DFT) calculation which is extremely computational costly. An effective solution is to incorporate the unlabeled molecules in a semi-supervised fashion. However, learning semi-supervised representation for large amounts of molecules is challenging, including the joint representation issue of both molecular essence and structure, the conflict between representation and property leaning. Here we propose a novel framework called Active Semi-supervised Graph Neural Network (ASGN) by incorporating both labeled and unlabeled molecules. Specifically, ASGN adopts a teacher-student framework. In the teacher model, we propose a novel semi-supervised learning method to learn general representation that jointly exploits information from molecular structure and molecular distribution. Then in the student model, we target at property prediction task to deal with the learning loss conflict. At last, we proposed a novel active learning strategy in terms of molecular diversities to select informative data during the whole framework learning. We conduct extensive experiments on several public datasets. Experimental results show the remarkable performance of our ASGN framework.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge