Chandravardhan Singh Raghaw
A multi-temporal multi-spectral attention-augmented deep convolution neural network with contrastive learning for crop yield prediction
Sep 19, 2025



Abstract:Precise yield prediction is essential for agricultural sustainability and food security. However, climate change complicates accurate yield prediction by affecting major factors such as weather conditions, soil fertility, and farm management systems. Advances in technology have played an essential role in overcoming these challenges by leveraging satellite monitoring and data analysis for precise yield estimation. Current methods rely on spatio-temporal data for predicting crop yield, but they often struggle with multi-spectral data, which is crucial for evaluating crop health and growth patterns. To resolve this challenge, we propose a novel Multi-Temporal Multi-Spectral Yield Prediction Network, MTMS-YieldNet, that integrates spectral data with spatio-temporal information to effectively capture the correlations and dependencies between them. While existing methods that rely on pre-trained models trained on general visual data, MTMS-YieldNet utilizes contrastive learning for feature discrimination during pre-training, focusing on capturing spatial-spectral patterns and spatio-temporal dependencies from remote sensing data. Both quantitative and qualitative assessments highlight the excellence of the proposed MTMS-YieldNet over seven existing state-of-the-art methods. MTMS-YieldNet achieves MAPE scores of 0.336 on Sentinel-1, 0.353 on Landsat-8, and an outstanding 0.331 on Sentinel-2, demonstrating effective yield prediction performance across diverse climatic and seasonal conditions. The outstanding performance of MTMS-YieldNet improves yield predictions and provides valuable insights that can assist farmers in making better decisions, potentially improving crop yields.
MNet-SAt: A Multiscale Network with Spatial-enhanced Attention for Segmentation of Polyps in Colonoscopy
Dec 27, 2024Abstract:Objective: To develop a novel deep learning framework for the automated segmentation of colonic polyps in colonoscopy images, overcoming the limitations of current approaches in preserving precise polyp boundaries, incorporating multi-scale features, and modeling spatial dependencies that accurately reflect the intricate and diverse morphology of polyps. Methods: To address these limitations, we propose a novel Multiscale Network with Spatial-enhanced Attention (MNet-SAt) for polyp segmentation in colonoscopy images. This framework incorporates four key modules: Edge-Guided Feature Enrichment (EGFE) preserves edge information for improved boundary quality; Multi-Scale Feature Aggregator (MSFA) extracts and aggregates multi-scale features across channel spatial dimensions, focusing on salient regions; Spatial-Enhanced Attention (SEAt) captures spatial-aware global dependencies within the multi-scale aggregated features, emphasizing the region of interest; and Channel-Enhanced Atrous Spatial Pyramid Pooling (CE-ASPP) resamples and recalibrates attentive features across scales. Results: We evaluated MNet-SAt on the Kvasir-SEG and CVC-ClinicDB datasets, achieving Dice Similarity Coefficients of 96.61% and 98.60%, respectively. Conclusion: Both quantitative (DSC) and qualitative assessments highlight MNet-SAt's superior performance and generalization capabilities compared to existing methods. Significance: MNet-SAt's high accuracy in polyp segmentation holds promise for improving clinical workflows in early polyp detection and more effective treatment, contributing to reduced colorectal cancer mortality rates.
Sentiment and Hashtag-aware Attentive Deep Neural Network for Multimodal Post Popularity Prediction
Dec 14, 2024Abstract:Social media users articulate their opinions on a broad spectrum of subjects and share their experiences through posts comprising multiple modes of expression, leading to a notable surge in such multimodal content on social media platforms. Nonetheless, accurately forecasting the popularity of these posts presents a considerable challenge. Prevailing methodologies primarily center on the content itself, thereby overlooking the wealth of information encapsulated within alternative modalities such as visual demographics, sentiments conveyed through hashtags and adequately modeling the intricate relationships among hashtags, texts, and accompanying images. This oversight limits the ability to capture emotional connection and audience relevance, significantly influencing post popularity. To address these limitations, we propose a seNtiment and hAshtag-aware attentive deep neuRal netwoRk for multimodAl posT pOpularity pRediction, herein referred to as NARRATOR that extracts visual demographics from faces appearing in images and discerns sentiment from hashtag usage, providing a more comprehensive understanding of the factors influencing post popularity Moreover, we introduce a hashtag-guided attention mechanism that leverages hashtags as navigational cues, guiding the models focus toward the most pertinent features of textual and visual modalities, thus aligning with target audience interests and broader social media context. Experimental results demonstrate that NARRATOR outperforms existing methods by a significant margin on two real-world datasets. Furthermore, ablation studies underscore the efficacy of integrating visual demographics, sentiment analysis of hashtags, and hashtag-guided attention mechanisms in enhancing the performance of post popularity prediction, thereby facilitating increased audience relevance, emotional engagement, and aesthetic appeal.
An Explainable Contrastive-based Dilated Convolutional Network with Transformer for Pediatric Pneumonia Detection
Oct 21, 2024



Abstract:Pediatric pneumonia remains a significant global threat, posing a larger mortality risk than any other communicable disease. According to UNICEF, it is a leading cause of mortality in children under five and requires prompt diagnosis. Early diagnosis using chest radiographs is the prevalent standard, but limitations include low radiation levels in unprocessed images and data imbalance issues. This necessitates the development of efficient, computer-aided diagnosis techniques. To this end, we propose a novel EXplainable Contrastive-based Dilated Convolutional Network with Transformer (XCCNet) for pediatric pneumonia detection. XCCNet harnesses the spatial power of dilated convolutions and the global insights from contrastive-based transformers for effective feature refinement. A robust chest X-ray processing module tackles low-intensity radiographs, while adversarial-based data augmentation mitigates the skewed distribution of chest X-rays in the dataset. Furthermore, we actively integrate an explainability approach through feature visualization, directly aligning it with the attention region that pinpoints the presence of pneumonia or normality in radiographs. The efficacy of XCCNet is comprehensively assessed on four publicly available datasets. Extensive performance evaluation demonstrates the superiority of XCCNet compared to state-of-the-art methods.
A Hybrid Filtering for Micro-video Hashtag Recommendation using Graph-based Deep Neural Network
Oct 14, 2024


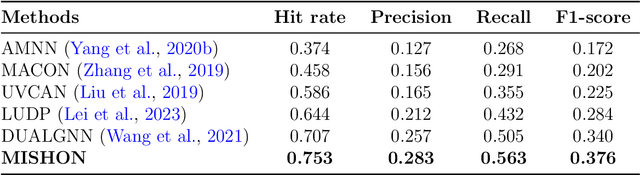
Abstract:Due to the growing volume of user generated content, hashtags are employed as topic indicators to manage content efficiently on social media platforms. However, finding these vital topics is challenging in microvideos since they contain substantial information in a short duration. Existing methods that recommend hashtags for microvideos primarily focus on content and personalization while disregarding relatedness among users. Moreover, the cold start user issue prevails in hashtag recommendation systems. Considering the above, we propose a hybrid filtering based MIcro-video haSHtag recommendatiON MISHON technique to recommend hashtags for micro-videos. Besides content based filtering, we employ user-based collaborative filtering to enhance recommendations. Since hashtags reflect users topical interests, we find similar users based on historical tagging behavior to model user relatedness. We employ a graph-based deep neural network to model user to user, modality to modality, and user to modality interactions. We then use refined modality specific and user representations to recommend pertinent hashtags for microvideos. The empirical results on three real world datasets demonstrate that MISHON attains a comparative enhancement of 3.6, 2.8, and 6.5 reported in percentage concerning the F1 score, respectively. Since cold start users exist whose historical tagging information is unavailable, we also propose a content and social influence based technique to model the relatedness of cold start users with influential users. The proposed solution shows a relative improvement of 15.8 percent in the F1 score over its content only counterpart. These results show that the proposed framework mitigates the cold start user problem.
CoTCoNet: An Optimized Coupled Transformer-Convolutional Network with an Adaptive Graph Reconstruction for Leukemia Detection
Oct 11, 2024



Abstract:Swift and accurate blood smear analysis is an effective diagnostic method for leukemia and other hematological malignancies. However, manual leukocyte count and morphological evaluation using a microscope is time-consuming and prone to errors. Conventional image processing methods also exhibit limitations in differentiating cells due to the visual similarity between malignant and benign cell morphology. This limitation is further compounded by the skewed training data that hinders the extraction of reliable and pertinent features. In response to these challenges, we propose an optimized Coupled Transformer Convolutional Network (CoTCoNet) framework for the classification of leukemia, which employs a well-designed transformer integrated with a deep convolutional network to effectively capture comprehensive global features and scalable spatial patterns, enabling the identification of complex and large-scale hematological features. Further, the framework incorporates a graph-based feature reconstruction module to reveal the hidden or unobserved hard-to-see biological features of leukocyte cells and employs a Population-based Meta-Heuristic Algorithm for feature selection and optimization. To mitigate data imbalance issues, we employ a synthetic leukocyte generator. In the evaluation phase, we initially assess CoTCoNet on a dataset containing 16,982 annotated cells, and it achieves remarkable accuracy and F1-Score rates of 0.9894 and 0.9893, respectively. To broaden the generalizability of our model, we evaluate it across four publicly available diverse datasets, which include the aforementioned dataset. This evaluation demonstrates that our method outperforms current state-of-the-art approaches. We also incorporate an explainability approach in the form of feature visualization closely aligned with cell annotations to provide a deeper understanding of the framework.
A Comprehensive Survey of Mamba Architectures for Medical Image Analysis: Classification, Segmentation, Restoration and Beyond
Oct 03, 2024Abstract:Mamba, a special case of the State Space Model, is gaining popularity as an alternative to template-based deep learning approaches in medical image analysis. While transformers are powerful architectures, they have drawbacks, including quadratic computational complexity and an inability to address long-range dependencies efficiently. This limitation affects the analysis of large and complex datasets in medical imaging, where there are many spatial and temporal relationships. In contrast, Mamba offers benefits that make it well-suited for medical image analysis. It has linear time complexity, which is a significant improvement over transformers. Mamba processes longer sequences without attention mechanisms, enabling faster inference and requiring less memory. Mamba also demonstrates strong performance in merging multimodal data, improving diagnosis accuracy and patient outcomes. The organization of this paper allows readers to appreciate the capabilities of Mamba in medical imaging step by step. We begin by defining core concepts of SSMs and models, including S4, S5, and S6, followed by an exploration of Mamba architectures such as pure Mamba, U-Net variants, and hybrid models with convolutional neural networks, transformers, and Graph Neural Networks. We also cover Mamba optimizations, techniques and adaptations, scanning, datasets, applications, experimental results, and conclude with its challenges and future directions in medical imaging. This review aims to demonstrate the transformative potential of Mamba in overcoming existing barriers within medical imaging while paving the way for innovative advancements in the field. A comprehensive list of Mamba architectures applied in the medical field, reviewed in this work, is available at Github.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge