Bradley Taylor
PRISM: Patient Records Interpretation for Semantic Clinical Trial Matching using Large Language Models
Apr 27, 2024



Abstract:Clinical trial matching is the task of identifying trials for which patients may be potentially eligible. Typically, this task is labor-intensive and requires detailed verification of patient electronic health records (EHRs) against the stringent inclusion and exclusion criteria of clinical trials. This process is manual, time-intensive, and challenging to scale up, resulting in many patients missing out on potential therapeutic options. Recent advancements in Large Language Models (LLMs) have made automating patient-trial matching possible, as shown in multiple concurrent research studies. However, the current approaches are confined to constrained, often synthetic datasets that do not adequately mirror the complexities encountered in real-world medical data. In this study, we present the first, end-to-end large-scale empirical evaluation of clinical trial matching using real-world EHRs. Our study showcases the capability of LLMs to accurately match patients with appropriate clinical trials. We perform experiments with proprietary LLMs, including GPT-4 and GPT-3.5, as well as our custom fine-tuned model called OncoLLM and show that OncoLLM, despite its significantly smaller size, not only outperforms GPT-3.5 but also matches the performance of qualified medical doctors. All experiments were carried out on real-world EHRs that include clinical notes and available clinical trials from a single cancer center in the United States.
Onco-Retriever: Generative Classifier for Retrieval of EHR Records in Oncology
Apr 10, 2024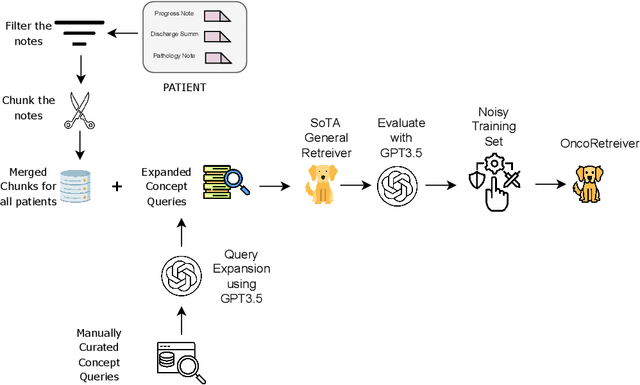
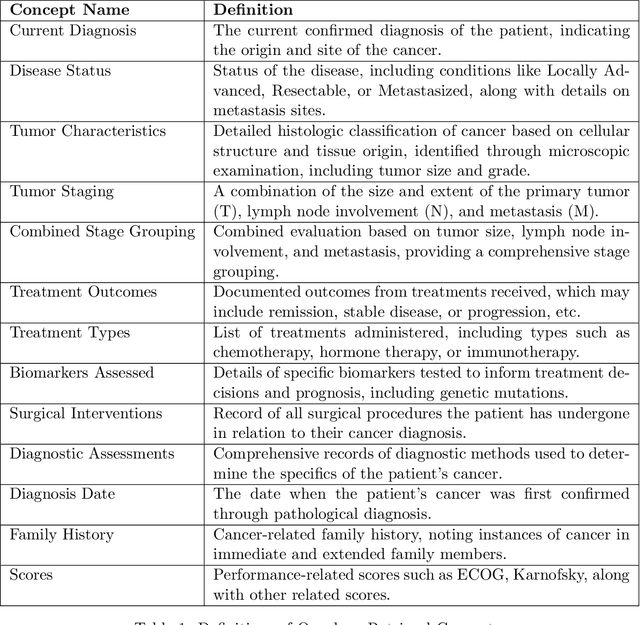
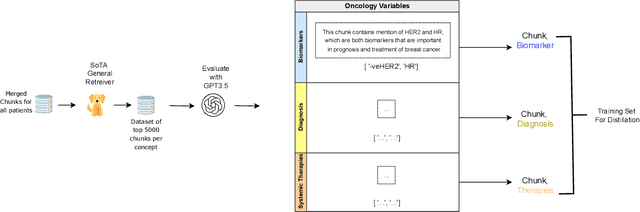
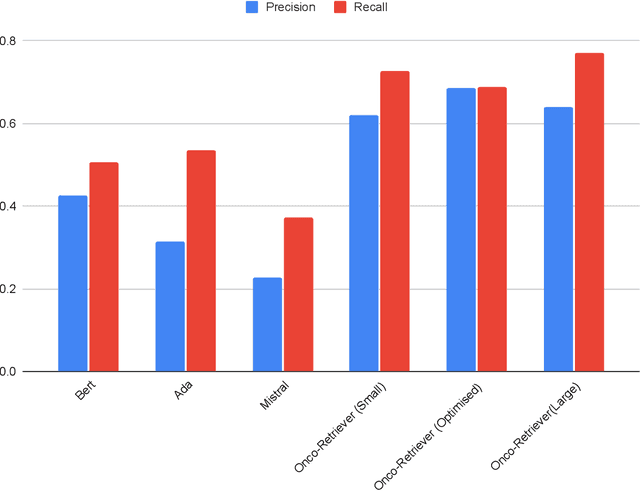
Abstract:Retrieving information from EHR systems is essential for answering specific questions about patient journeys and improving the delivery of clinical care. Despite this fact, most EHR systems still rely on keyword-based searches. With the advent of generative large language models (LLMs), retrieving information can lead to better search and summarization capabilities. Such retrievers can also feed Retrieval-augmented generation (RAG) pipelines to answer any query. However, the task of retrieving information from EHR real-world clinical data contained within EHR systems in order to solve several downstream use cases is challenging due to the difficulty in creating query-document support pairs. We provide a blueprint for creating such datasets in an affordable manner using large language models. Our method results in a retriever that is 30-50 F-1 points better than propriety counterparts such as Ada and Mistral for oncology data elements. We further compare our model, called Onco-Retriever, against fine-tuned PubMedBERT model as well. We conduct an extensive manual evaluation on real-world EHR data along with latency analysis of the different models and provide a path forward for healthcare organizations to build domain-specific retrievers.
The NLP Sandbox: an efficient model-to-data system to enable federated and unbiased evaluation of clinical NLP models
Jun 28, 2022



Abstract:Objective The evaluation of natural language processing (NLP) models for clinical text de-identification relies on the availability of clinical notes, which is often restricted due to privacy concerns. The NLP Sandbox is an approach for alleviating the lack of data and evaluation frameworks for NLP models by adopting a federated, model-to-data approach. This enables unbiased federated model evaluation without the need for sharing sensitive data from multiple institutions. Materials and Methods We leveraged the Synapse collaborative framework, containerization software, and OpenAPI generator to build the NLP Sandbox (nlpsandbox.io). We evaluated two state-of-the-art NLP de-identification focused annotation models, Philter and NeuroNER, using data from three institutions. We further validated model performance using data from an external validation site. Results We demonstrated the usefulness of the NLP Sandbox through de-identification clinical model evaluation. The external developer was able to incorporate their model into the NLP Sandbox template and provide user experience feedback. Discussion We demonstrated the feasibility of using the NLP Sandbox to conduct a multi-site evaluation of clinical text de-identification models without the sharing of data. Standardized model and data schemas enable smooth model transfer and implementation. To generalize the NLP Sandbox, work is required on the part of data owners and model developers to develop suitable and standardized schemas and to adapt their data or model to fit the schemas. Conclusions The NLP Sandbox lowers the barrier to utilizing clinical data for NLP model evaluation and facilitates federated, multi-site, unbiased evaluation of NLP models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge