Bojian Yin
Traces Propagation: Memory-Efficient and Scalable Forward-Only Learning in Spiking Neural Networks
Sep 16, 2025Abstract:Spiking Neural Networks (SNNs) provide an efficient framework for processing dynamic spatio-temporal signals and for investigating the learning principles underlying biological neural systems. A key challenge in training SNNs is to solve both spatial and temporal credit assignment. The dominant approach for training SNNs is Backpropagation Through Time (BPTT) with surrogate gradients. However, BPTT is in stark contrast with the spatial and temporal locality observed in biological neural systems and leads to high computational and memory demands, limiting efficient training strategies and on-device learning. Although existing local learning rules achieve local temporal credit assignment by leveraging eligibility traces, they fail to address the spatial credit assignment without resorting to auxiliary layer-wise matrices, which increase memory overhead and hinder scalability, especially on embedded devices. In this work, we propose Traces Propagation (TP), a forward-only, memory-efficient, scalable, and fully local learning rule that combines eligibility traces with a layer-wise contrastive loss without requiring auxiliary layer-wise matrices. TP outperforms other fully local learning rules on NMNIST and SHD datasets. On more complex datasets such as DVS-GESTURE and DVS-CIFAR10, TP showcases competitive performance and scales effectively to deeper SNN architectures such as VGG-9, while providing favorable memory scaling compared to prior fully local scalable rules, for datasets with a significant number of classes. Finally, we show that TP is well suited for practical fine-tuning tasks, such as keyword spotting on the Google Speech Commands dataset, thus paving the way for efficient learning at the edge.
Stochastic Variational Propagation: Local, Scalable and Efficient Alternative to Backpropagation
May 08, 2025Abstract:Backpropagation (BP) is the cornerstone of deep learning, but its reliance on global gradient synchronization limits scalability and imposes significant memory overhead. We propose Stochastic Variational Propagation (SVP), a scalable alternative that reframes training as hierarchical variational inference. SVP treats layer activations as latent variables and optimizes local Evidence Lower Bounds (ELBOs), enabling independent, local updates while preserving global coherence. However, directly applying KL divergence in layer-wise ELBOs risks inter-layer's representation collapse due to excessive compression. To prevent this, SVP projects activations into low-dimensional spaces via fixed random matrices, ensuring information preservation and representational diversity. Combined with a feature alignment loss for inter-layer consistency, SVP achieves competitive accuracy with BP across diverse architectures (MLPs, CNNs, Transformers) and datasets (MNIST to ImageNet), reduces memory usage by up to 4x, and significantly improves scalability. More broadly, SVP introduces a probabilistic perspective to deep representation learning, opening pathways toward more modular and interpretable neural network design.
Never Reset Again: A Mathematical Framework for Continual Inference in Recurrent Neural Networks
Dec 20, 2024Abstract:Recurrent Neural Networks (RNNs) are widely used for sequential processing but face fundamental limitations with continual inference due to state saturation, requiring disruptive hidden state resets. However, reset-based methods impose synchronization requirements with input boundaries and increase computational costs at inference. To address this, we propose an adaptive loss function that eliminates the need for resets during inference while preserving high accuracy over extended sequences. By combining cross-entropy and Kullback-Leibler divergence, the loss dynamically modulates the gradient based on input informativeness, allowing the network to differentiate meaningful data from noise and maintain stable representations over time. Experimental results demonstrate that our reset-free approach outperforms traditional reset-based methods when applied to a variety of RNNs, particularly in continual tasks, enhancing both the theoretical and practical capabilities of RNNs for streaming applications.
Efficient Uncertainty Estimation in Spiking Neural Networks via MC-dropout
Apr 20, 2023Abstract:Spiking neural networks (SNNs) have gained attention as models of sparse and event-driven communication of biological neurons, and as such have shown increasing promise for energy-efficient applications in neuromorphic hardware. As with classical artificial neural networks (ANNs), predictive uncertainties are important for decision making in high-stakes applications, such as autonomous vehicles, medical diagnosis, and high frequency trading. Yet, discussion of uncertainty estimation in SNNs is limited, and approaches for uncertainty estimation in artificial neural networks (ANNs) are not directly applicable to SNNs. Here, we propose an efficient Monte Carlo(MC)-dropout based approach for uncertainty estimation in SNNs. Our approach exploits the time-step mechanism of SNNs to enable MC-dropout in a computationally efficient manner, without introducing significant overheads during training and inference while demonstrating high accuracy and uncertainty quality.
Accurate online training of dynamical spiking neural networks through Forward Propagation Through Time
Dec 20, 2021



Abstract:The event-driven and sparse nature of communication between spiking neurons in the brain holds great promise for flexible and energy-efficient AI. Recent advances in learning algorithms have demonstrated that recurrent networks of spiking neurons can be effectively trained to achieve competitive performance compared to standard recurrent neural networks. Still, as these learning algorithms use error-backpropagation through time (BPTT), they suffer from high memory requirements, are slow to train, and are incompatible with online learning. This limits the application of these learning algorithms to relatively small networks and to limited temporal sequence lengths. Online approximations to BPTT with lower computational and memory complexity have been proposed (e-prop, OSTL), but in practice also suffer from memory limitations and, as approximations, do not outperform standard BPTT training. Here, we show how a recently developed alternative to BPTT, Forward Propagation Through Time (FPTT) can be applied in spiking neural networks. Different from BPTT, FPTT attempts to minimize an ongoing dynamically regularized risk on the loss. As a result, FPTT can be computed in an online fashion and has fixed complexity with respect to the sequence length. When combined with a novel dynamic spiking neuron model, the Liquid-Time-Constant neuron, we show that SNNs trained with FPTT outperform online BPTT approximations, and approach or exceed offline BPTT accuracy on temporal classification tasks. This approach thus makes it feasible to train SNNs in a memory-friendly online fashion on long sequences and scale up SNNs to novel and complex neural architectures.
Accurate and efficient time-domain classification with adaptive spiking recurrent neural networks
Mar 12, 2021Abstract:Inspired by more detailed modeling of biological neurons, Spiking neural networks (SNNs) have been investigated both as more biologically plausible and potentially more powerful models of neural computation, and also with the aim of extracting biological neurons' energy efficiency; the performance of such networks however has remained lacking compared to classical artificial neural networks (ANNs). Here, we demonstrate how a novel surrogate gradient combined with recurrent networks of tunable and adaptive spiking neurons yields state-of-the-art for SNNs on challenging benchmarks in the time-domain, like speech and gesture recognition. This also exceeds the performance of standard classical recurrent neural networks (RNNs) and approaches that of the best modern ANNs. As these SNNs exhibit sparse spiking, we show that they theoretically are one to three orders of magnitude more computationally efficient compared to RNNs with comparable performance. Together, this positions SNNs as an attractive solution for AI hardware implementations.
Effective and Efficient Computation with Multiple-timescale Spiking Recurrent Neural Networks
Jun 16, 2020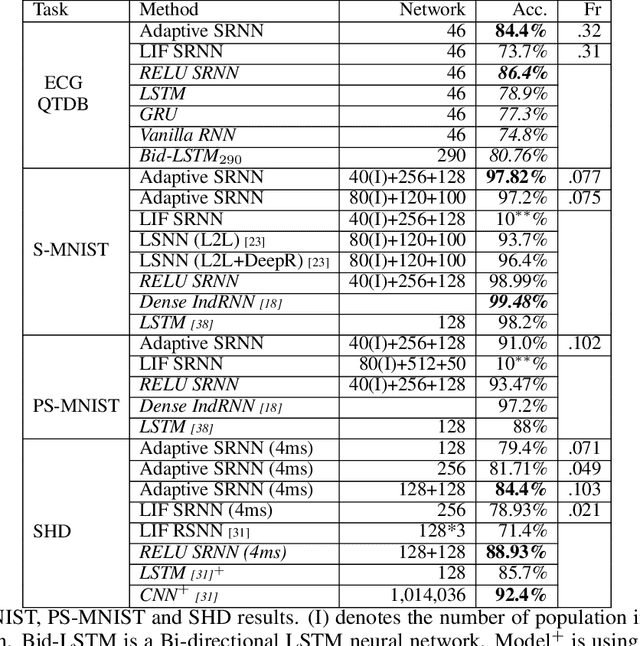
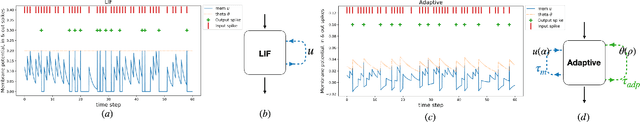
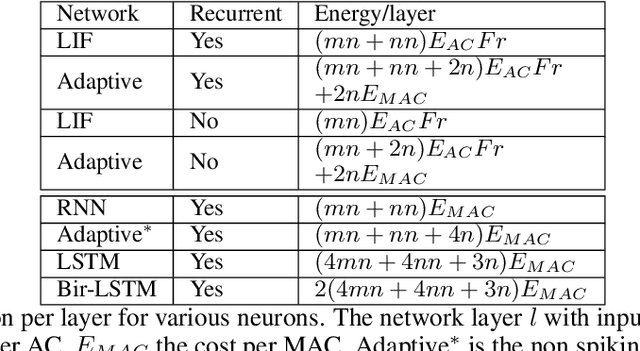
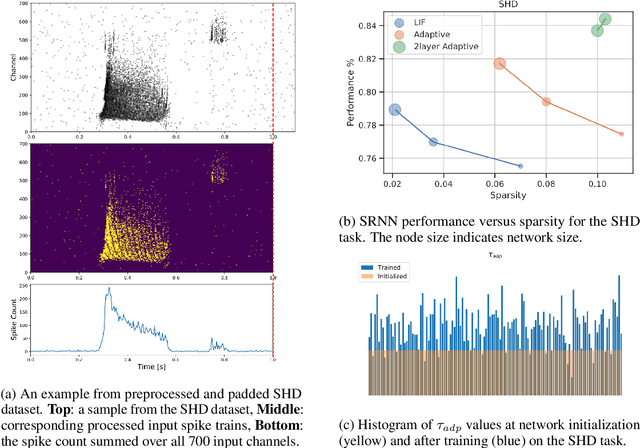
Abstract:The emergence of brain-inspired neuromorphic computing as a paradigm for edge AI is motivating the search for high-performance and efficient spiking neural networks to run on this hardware. However, compared to classical neural networks in deep learning, current spiking neural networks lack competitive performance in compelling areas. Here, for sequential and streaming tasks, we demonstrate how a novel type of adaptive spiking recurrent neural network (SRNN) is able to achieve state-of-the-art performance compared to other spiking neural networks and almost reach or exceed the performance of classical recurrent neural networks (RNNs) while exhibiting sparse activity. From this, we calculate a $>$100x energy improvement for our SRNNs over classical RNNs on the harder tasks. To achieve this, we model standard and adaptive multiple-timescale spiking neurons as self-recurrent neural units, and leverage surrogate gradients and auto-differentiation in the PyTorch Deep Learning framework to efficiently implement backpropagation-through-time, including learning of the important spiking neuron parameters to adapt our spiking neurons to the tasks.
LocalNorm: Robust Image Classification through Dynamically Regularized Normalization
Mar 04, 2019
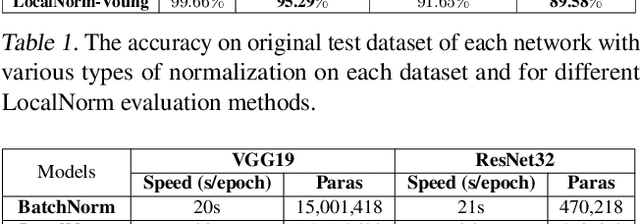

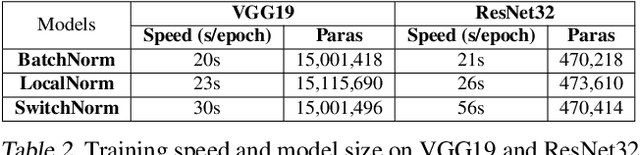
Abstract:While modern convolutional neural networks achieve outstanding accuracy on many image classification tasks, they are, compared to humans, much more sensitive to image degradation. Here, we describe a variant of Batch Normalization, LocalNorm, that regularizes the normalization layer in the spirit of Dropout while dynamically adapting to the local image intensity and contrast at test-time. We show that the resulting deep neural networks are much more resistant to noise-induced image degradation, improving accuracy by up to three times, while achieving the same or slightly better accuracy on non-degraded classical benchmarks. In computational terms, LocalNorm adds negligible training cost and little or no cost at inference time, and can be applied to already-trained networks in a straightforward manner.
An image representation based convolutional network for DNA classification
Jun 13, 2018



Abstract:The folding structure of the DNA molecule combined with helper molecules, also referred to as the chromatin, is highly relevant for the functional properties of DNA. The chromatin structure is largely determined by the underlying primary DNA sequence, though the interaction is not yet fully understood. In this paper we develop a convolutional neural network that takes an image-representation of primary DNA sequence as its input, and predicts key determinants of chromatin structure. The method is developed such that it is capable of detecting interactions between distal elements in the DNA sequence, which are known to be highly relevant. Our experiments show that the method outperforms several existing methods both in terms of prediction accuracy and training time.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge