Bogdan Gabrys
Machine Learning Methods for Small Data and Upstream Bioprocessing Applications: A Comprehensive Review
Jun 14, 2025Abstract:Data is crucial for machine learning (ML) applications, yet acquiring large datasets can be costly and time-consuming, especially in complex, resource-intensive fields like biopharmaceuticals. A key process in this industry is upstream bioprocessing, where living cells are cultivated and optimised to produce therapeutic proteins and biologics. The intricate nature of these processes, combined with high resource demands, often limits data collection, resulting in smaller datasets. This comprehensive review explores ML methods designed to address the challenges posed by small data and classifies them into a taxonomy to guide practical applications. Furthermore, each method in the taxonomy was thoroughly analysed, with a detailed discussion of its core concepts and an evaluation of its effectiveness in tackling small data challenges, as demonstrated by application results in the upstream bioprocessing and other related domains. By analysing how these methods tackle small data challenges from different perspectives, this review provides actionable insights, identifies current research gaps, and offers guidance for leveraging ML in data-constrained environments.
FactSelfCheck: Fact-Level Black-Box Hallucination Detection for LLMs
Mar 21, 2025Abstract:Large Language Models (LLMs) frequently generate hallucinated content, posing significant challenges for applications where factuality is crucial. While existing hallucination detection methods typically operate at the sentence level or passage level, we propose FactSelfCheck, a novel black-box sampling-based method that enables fine-grained fact-level detection. Our approach represents text as knowledge graphs consisting of facts in the form of triples. Through analyzing factual consistency across multiple LLM responses, we compute fine-grained hallucination scores without requiring external resources or training data. Our evaluation demonstrates that FactSelfCheck performs competitively with leading sampling-based methods while providing more detailed insights. Most notably, our fact-level approach significantly improves hallucination correction, achieving a 35% increase in factual content compared to the baseline, while sentence-level SelfCheckGPT yields only an 8% improvement. The granular nature of our detection enables more precise identification and correction of hallucinated content.
Hallucination Detection in LLMs Using Spectral Features of Attention Maps
Feb 24, 2025Abstract:Large Language Models (LLMs) have demonstrated remarkable performance across various tasks but remain prone to hallucinations. Detecting hallucinations is essential for safety-critical applications, and recent methods leverage attention map properties to this end, though their effectiveness remains limited. In this work, we investigate the spectral features of attention maps by interpreting them as adjacency matrices of graph structures. We propose the $\text{LapEigvals}$ method, which utilises the top-$k$ eigenvalues of the Laplacian matrix derived from the attention maps as an input to hallucination detection probes. Empirical evaluations demonstrate that our approach achieves state-of-the-art hallucination detection performance among attention-based methods. Extensive ablation studies further highlight the robustness and generalisation of $\text{LapEigvals}$, paving the way for future advancements in the hallucination detection domain.
Deep Reinforcement Learning for Digital Twin-Oriented Complex Networked Systems
Nov 09, 2024Abstract:The Digital Twin Oriented Complex Networked System (DT-CNS) aims to build and extend a Complex Networked System (CNS) model with progressively increasing dynamics complexity towards an accurate reflection of reality -- a Digital Twin of reality. Our previous work proposed evolutionary DT-CNSs to model the long-term adaptive network changes in an epidemic outbreak. This study extends this framework by proposeing the temporal DT-CNS model, where reinforcement learning-driven nodes make decisions on temporal directed interactions in an epidemic outbreak. We consider cooperative nodes, as well as egocentric and ignorant "free-riders" in the cooperation. We describe this epidemic spreading process with the Susceptible-Infected-Recovered ($SIR$) model and investigate the impact of epidemic severity on the epidemic resilience for different types of nodes. Our experimental results show that (i) the full cooperation leads to a higher reward and lower infection number than a cooperation with egocentric or ignorant "free-riders"; (ii) an increasing number of "free-riders" in a cooperation leads to a smaller reward, while an increasing number of egocentric "free-riders" further escalate the infection numbers and (iii) higher infection rates and a slower recovery weakens networks' resilience to severe epidemic outbreaks. These findings also indicate that promoting cooperation and reducing "free-riders" can improve public health during epidemics.
Hyperbox Mixture Regression for Process Performance Prediction in Antibody Production
Nov 03, 2024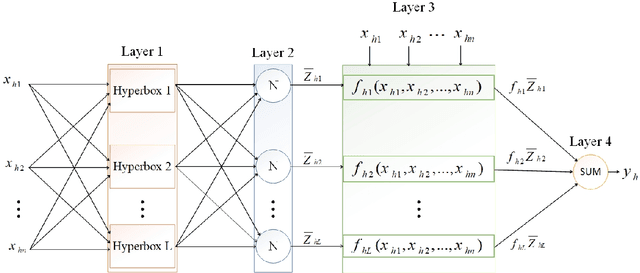
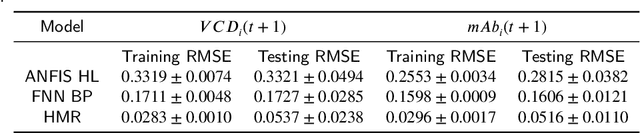
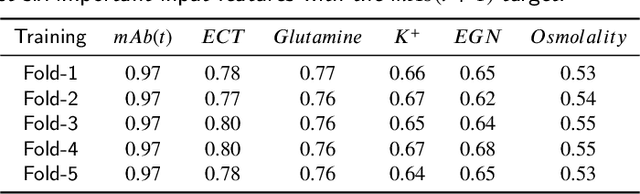
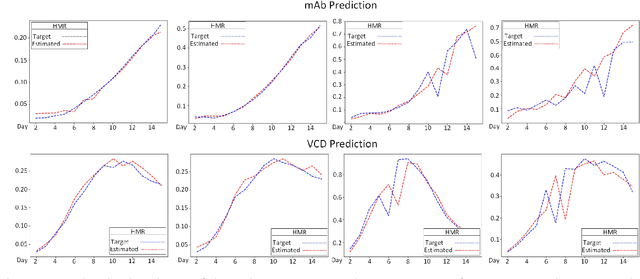
Abstract:This paper addresses the challenges of predicting bioprocess performance, particularly in monoclonal antibody (mAb) production, where conventional statistical methods often fall short due to time-series data's complexity and high dimensionality. We propose a novel Hyperbox Mixture Regression (HMR) model which employs hyperbox-based input space partitioning to enhance predictive accuracy while managing uncertainty inherent in bioprocess data. The HMR model is designed to dynamically generate hyperboxes for input samples in a single-pass process, thereby improving learning speed and reducing computational complexity. Our experimental study utilizes a dataset that contains 106 bioreactors. This study evaluates the model's performance in predicting critical quality attributes in monoclonal antibody manufacturing over a 15-day cultivation period. The results demonstrate that the HMR model outperforms comparable approximators in accuracy and learning speed and maintains interpretability and robustness under uncertain conditions. These findings underscore the potential of HMR as a powerful tool for enhancing predictive analytics in bioprocessing applications.
Uncertainty Quantification Using Ensemble Learning and Monte Carlo Sampling for Performance Prediction and Monitoring in Cell Culture Processes
Sep 03, 2024



Abstract:Biopharmaceutical products, particularly monoclonal antibodies (mAbs), have gained prominence in the pharmaceutical market due to their high specificity and efficacy. As these products are projected to constitute a substantial portion of global pharmaceutical sales, the application of machine learning models in mAb development and manufacturing is gaining momentum. This paper addresses the critical need for uncertainty quantification in machine learning predictions, particularly in scenarios with limited training data. Leveraging ensemble learning and Monte Carlo simulations, our proposed method generates additional input samples to enhance the robustness of the model in small training datasets. We evaluate the efficacy of our approach through two case studies: predicting antibody concentrations in advance and real-time monitoring of glucose concentrations during bioreactor runs using Raman spectra data. Our findings demonstrate the effectiveness of the proposed method in estimating the uncertainty levels associated with process performance predictions and facilitating real-time decision-making in biopharmaceutical manufacturing. This contribution not only introduces a novel approach for uncertainty quantification but also provides insights into overcoming challenges posed by small training datasets in bioprocess development. The evaluation demonstrates the effectiveness of our method in addressing key challenges related to uncertainty estimation within upstream cell cultivation, illustrating its potential impact on enhancing process control and product quality in the dynamic field of biopharmaceuticals.
Transfer Learning and the Early Estimation of Single-Photon Source Quality using Machine Learning Methods
Aug 21, 2024Abstract:The use of single-photon sources (SPSs) is central to numerous systems and devices proposed amidst a modern surge in quantum technology. However, manufacturing schemes remain imperfect, and single-photon emission purity must often be experimentally verified via interferometry. Such a process is typically slow and costly, which has motivated growing research into whether SPS quality can be more rapidly inferred from incomplete emission statistics. Hence, this study is a sequel to previous work that demonstrated significant uncertainty in the standard method of quality estimation, i.e. the least-squares fitting of a physically motivated function, and asks: can machine learning (ML) do better? The study leverages eight datasets obtained from measurements involving an exemplary quantum emitter, i.e. a single InGaAs/GaAs epitaxial quantum dot; these eight contexts predominantly vary in the intensity of the exciting laser. Specifically, via a form of `transfer learning', five ML models, three linear and two ensemble-based, are trained on data from seven of the contexts and tested on the eighth. Validation metrics quickly reveal that even a linear regressor can outperform standard fitting when it is tested on the same contexts it was trained on, but the success of transfer learning is less assured, even though statistical analysis, made possible by data augmentation, suggests its superiority as an early estimator. Accordingly, the study concludes by discussing future strategies for grappling with the problem of SPS context dissimilarity, e.g. feature engineering and model adaptation.
Applications of Machine Learning in Biopharmaceutical Process Development and Manufacturing: Current Trends, Challenges, and Opportunities
Oct 16, 2023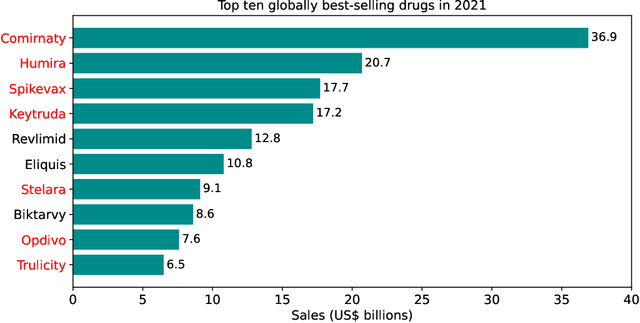

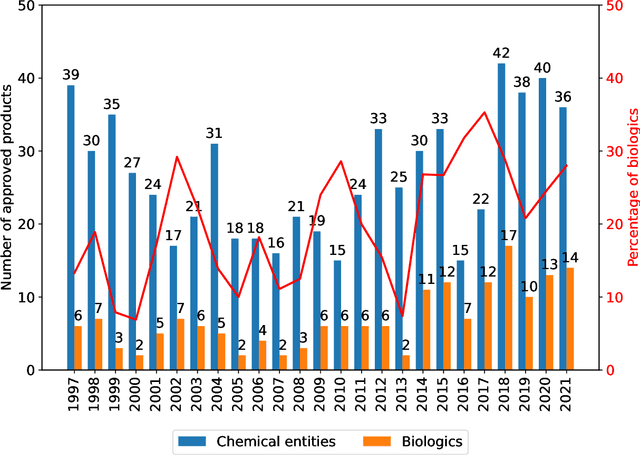
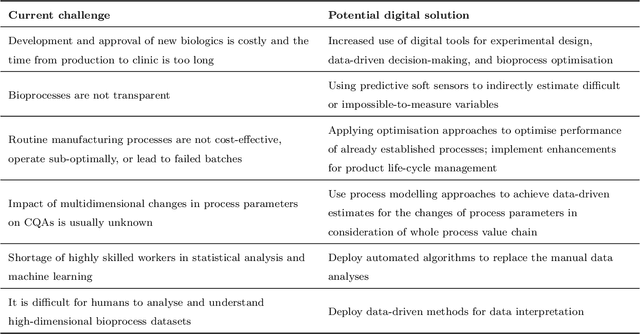
Abstract:While machine learning (ML) has made significant contributions to the biopharmaceutical field, its applications are still in the early stages in terms of providing direct support for quality-by-design based development and manufacturing of biopharmaceuticals, hindering the enormous potential for bioprocesses automation from their development to manufacturing. However, the adoption of ML-based models instead of conventional multivariate data analysis methods is significantly increasing due to the accumulation of large-scale production data. This trend is primarily driven by the real-time monitoring of process variables and quality attributes of biopharmaceutical products through the implementation of advanced process analytical technologies. Given the complexity and multidimensionality of a bioproduct design, bioprocess development, and product manufacturing data, ML-based approaches are increasingly being employed to achieve accurate, flexible, and high-performing predictive models to address the problems of analytics, monitoring, and control within the biopharma field. This paper aims to provide a comprehensive review of the current applications of ML solutions in a bioproduct design, monitoring, control, and optimisation of upstream, downstream, and product formulation processes. Finally, this paper thoroughly discusses the main challenges related to the bioprocesses themselves, process data, and the use of machine learning models in biopharmaceutical process development and manufacturing. Moreover, it offers further insights into the adoption of innovative machine learning methods and novel trends in the development of new digital biopharma solutions.
Heterogeneous Feature Representation for Digital Twin-Oriented Complex Networked Systems
Sep 23, 2023
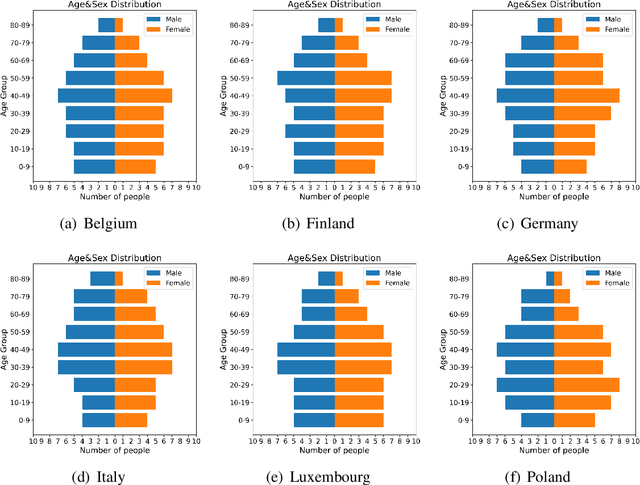
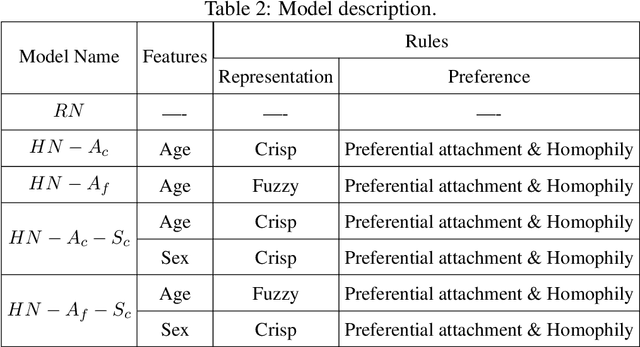
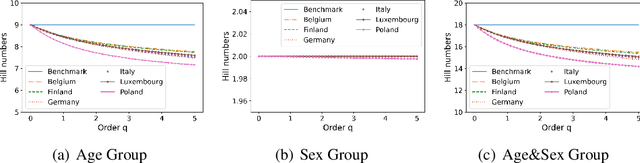
Abstract:Building models of Complex Networked Systems (CNS) that can accurately represent reality forms an important research area. To be able to reflect real world systems, the modelling needs to consider not only the intensity of interactions between the entities but also features of all the elements of the system. This study aims to improve the expressive power of node features in Digital Twin-Oriented Complex Networked Systems (DT-CNSs) with heterogeneous feature representation principles. This involves representing features with crisp feature values and fuzzy sets, each describing the objective and the subjective inductions of the nodes' features and feature differences. Our empirical analysis builds DT-CNSs to recreate realistic physical contact networks in different countries from real node feature distributions based on various representation principles and an optimised feature preference. We also investigate their respective disaster resilience to an epidemic outbreak starting from the most popular node. The results suggest that the increasing flexibility of feature representation with fuzzy sets improves the expressive power and enables more accurate modelling. In addition, the heterogeneous features influence the network structure and the speed of the epidemic outbreak, requiring various mitigation policies targeted at different people.
Digital Twin-Oriented Complex Networked Systems based on Heterogeneous node features and interaction rules
Aug 18, 2023Abstract:This study proposes an extendable modelling framework for Digital Twin-Oriented Complex Networked Systems (DT-CNSs) with a goal of generating networks that faithfully represent real systems. Modelling process focuses on (i) features of nodes and (ii) interaction rules for creating connections that are built based on individual node's preferences. We conduct experiments on simulation-based DT-CNSs that incorporate various features and rules about network growth and different transmissibilities related to an epidemic spread on these networks. We present a case study on disaster resilience of social networks given an epidemic outbreak by investigating the infection occurrence within specific time and social distance. The experimental results show how different levels of the structural and dynamics complexities, concerned with feature diversity and flexibility of interaction rules respectively, influence network growth and epidemic spread. The analysis revealed that, to achieve maximum disaster resilience, mitigation policies should be targeted at nodes with preferred features as they have higher infection risks and should be the focus of the epidemic control.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge