Baojuan Li
Learning Multitask Gaussian Bayesian Networks
May 11, 2022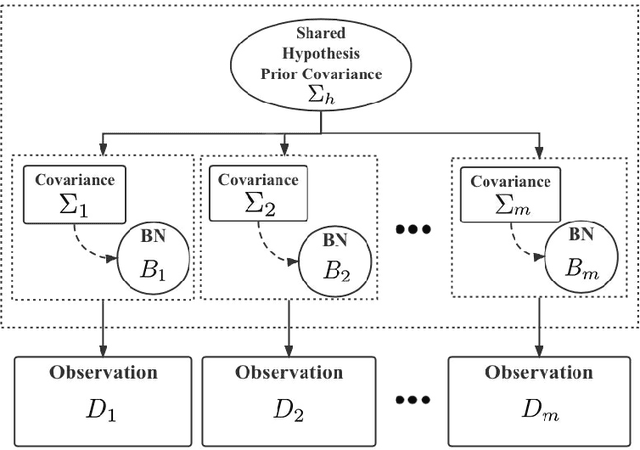
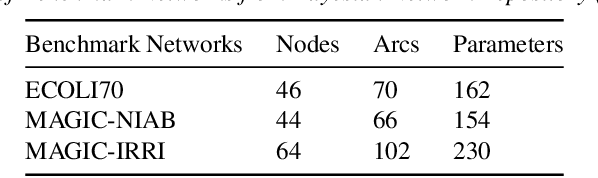
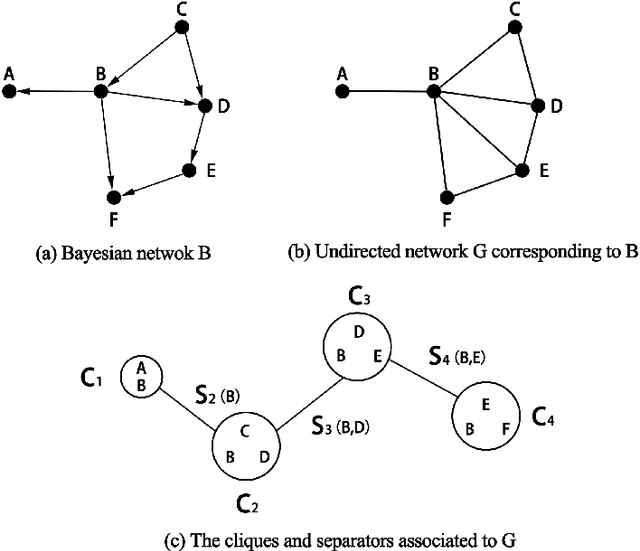
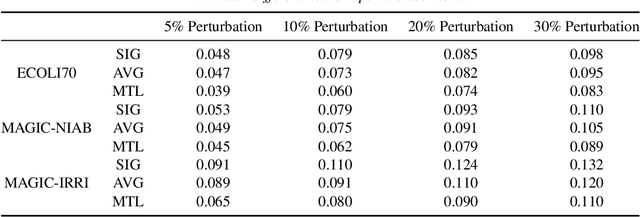
Abstract:Major depressive disorder (MDD) requires study of brain functional connectivity alterations for patients, which can be uncovered by resting-state functional magnetic resonance imaging (rs-fMRI) data. We consider the problem of identifying alterations of brain functional connectivity for a single MDD patient. This is particularly difficult since the amount of data collected during an fMRI scan is too limited to provide sufficient information for individual analysis. Additionally, rs-fMRI data usually has the characteristics of incompleteness, sparsity, variability, high dimensionality and high noise. To address these problems, we proposed a multitask Gaussian Bayesian network (MTGBN) framework capable for identifying individual disease-induced alterations for MDD patients. We assume that such disease-induced alterations show some degrees of similarity with the tool to learn such network structures from observations to understanding of how system are structured jointly from related tasks. First, we treat each patient in a class of observation as a task and then learn the Gaussian Bayesian networks (GBNs) of this data class by learning from all tasks that share a default covariance matrix that encodes prior knowledge. This setting can help us to learn more information from limited data. Next, we derive a closed-form formula of the complete likelihood function and use the Monte-Carlo Expectation-Maximization(MCEM) algorithm to search for the approximately best Bayesian network structures efficiently. Finally, we assess the performance of our methods with simulated and real-world rs-fMRI data.
BrainIB: Interpretable Brain Network-based Psychiatric Diagnosis with Graph Information Bottleneck
May 07, 2022



Abstract:Developing a new diagnostic models based on the underlying biological mechanisms rather than subjective symptoms for psychiatric disorders is an emerging consensus. Recently, machine learning-based classifiers using functional connectivity (FC) for psychiatric disorders and healthy controls are developed to identify brain markers. However, existing machine learningbased diagnostic models are prone to over-fitting (due to insufficient training samples) and perform poorly in new test environment. Furthermore, it is difficult to obtain explainable and reliable brain biomarkers elucidating the underlying diagnostic decisions. These issues hinder their possible clinical applications. In this work, we propose BrainIB, a new graph neural network (GNN) framework to analyze functional magnetic resonance images (fMRI), by leveraging the famed Information Bottleneck (IB) principle. BrainIB is able to identify the most informative regions in the brain (i.e., subgraph) and generalizes well to unseen data. We evaluate the performance of BrainIB against 6 popular brain network classification methods on two multi-site, largescale datasets and observe that our BrainIB always achieves the highest diagnosis accuracy. It also discovers the subgraph biomarkers which are consistent to clinical and neuroimaging findings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge