Huaning Wang
Learning Personalized Brain Functional Connectivity of MDD Patients from Multiple Sites via Federated Bayesian Networks
Jan 06, 2023Abstract:Identifying functional connectivity biomarkers of major depressive disorder (MDD) patients is essential to advance understanding of the disorder mechanisms and early intervention. However, due to the small sample size and the high dimension of available neuroimaging data, the performance of existing methods is often limited. Multi-site data could enhance the statistical power and sample size, while they are often subject to inter-site heterogeneity and data-sharing policies. In this paper, we propose a federated joint estimator, NOTEARS-PFL, for simultaneous learning of multiple Bayesian networks (BNs) with continuous optimization, to identify disease-induced alterations in MDD patients. We incorporate information shared between sites and site-specific information into the proposed federated learning framework to learn personalized BN structures by introducing the group fused lasso penalty. We develop the alternating direction method of multipliers, where in the local update step, the neuroimaging data is processed at each local site. Then the learned network structures are transmitted to the center for the global update. In particular, we derive a closed-form expression for the local update step and use the iterative proximal projection method to deal with the group fused lasso penalty in the global update step. We evaluate the performance of the proposed method on both synthetic and real-world multi-site rs-fMRI datasets. The results suggest that the proposed NOTEARS-PFL yields superior effectiveness and accuracy than the comparable methods.
Learning Multitask Gaussian Bayesian Networks
May 11, 2022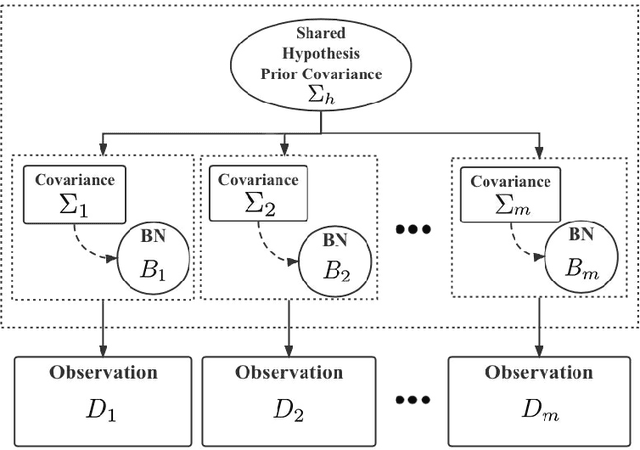
Abstract:Major depressive disorder (MDD) requires study of brain functional connectivity alterations for patients, which can be uncovered by resting-state functional magnetic resonance imaging (rs-fMRI) data. We consider the problem of identifying alterations of brain functional connectivity for a single MDD patient. This is particularly difficult since the amount of data collected during an fMRI scan is too limited to provide sufficient information for individual analysis. Additionally, rs-fMRI data usually has the characteristics of incompleteness, sparsity, variability, high dimensionality and high noise. To address these problems, we proposed a multitask Gaussian Bayesian network (MTGBN) framework capable for identifying individual disease-induced alterations for MDD patients. We assume that such disease-induced alterations show some degrees of similarity with the tool to learn such network structures from observations to understanding of how system are structured jointly from related tasks. First, we treat each patient in a class of observation as a task and then learn the Gaussian Bayesian networks (GBNs) of this data class by learning from all tasks that share a default covariance matrix that encodes prior knowledge. This setting can help us to learn more information from limited data. Next, we derive a closed-form formula of the complete likelihood function and use the Monte-Carlo Expectation-Maximization(MCEM) algorithm to search for the approximately best Bayesian network structures efficiently. Finally, we assess the performance of our methods with simulated and real-world rs-fMRI data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge