Bahareh Behboodi
RESECT-SEG: Open access annotations of intra-operative brain tumor ultrasound images
Jul 13, 2022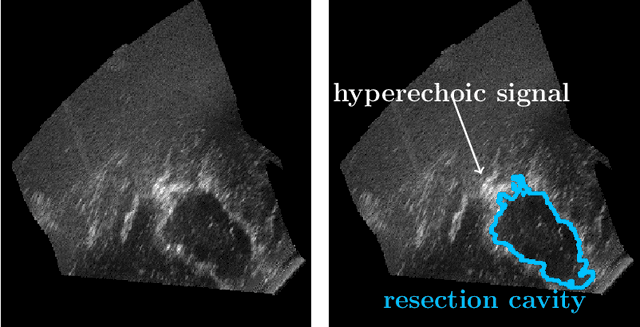
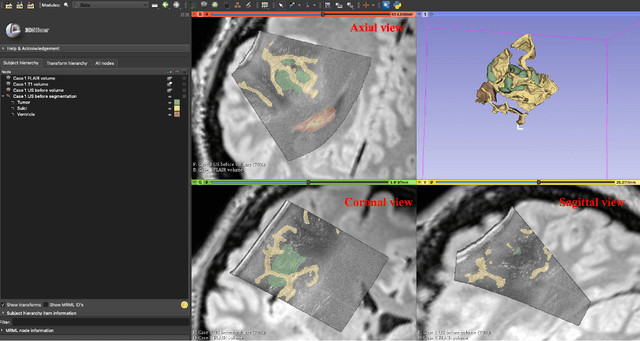
Abstract:Purpose: Registration and segmentation of magnetic resonance (MR) and ultrasound (US) images play an essential role in surgical planning and resection of brain tumors. However, validating these techniques is challenging due to the scarcity of publicly accessible sources with high-quality ground truth information. To this end, we propose a unique annotation dataset of tumor tissues and resection cavities from the previously published RESECT dataset (Xiao et al. 2017) to encourage a more rigorous assessments of image processing techniques. Acquisition and validation methods: The RESECT database consists of MR and intraoperative US (iUS) images of 23 patients who underwent resection surgeries. The proposed dataset contains tumor tissues and resection cavity annotations of the iUS images. The quality of annotations were validated by two highly experienced neurosurgeons through several assessment criteria. Data format and availability: Annotations of tumor tissues and resection cavities are provided in 3D NIFTI formats. Both sets of annotations are accessible online in the \url{https://osf.io/6y4db}. Discussion and potential applications: The proposed database includes tumor tissue and resection cavity annotations from real-world clinical ultrasound brain images to evaluate segmentation and registration methods. These labels could also be used to train deep learning approaches. Eventually, this dataset should further improve the quality of image guidance in neurosurgery.
Automatic 3D Ultrasound Segmentation of Uterus Using Deep Learning
Sep 20, 2021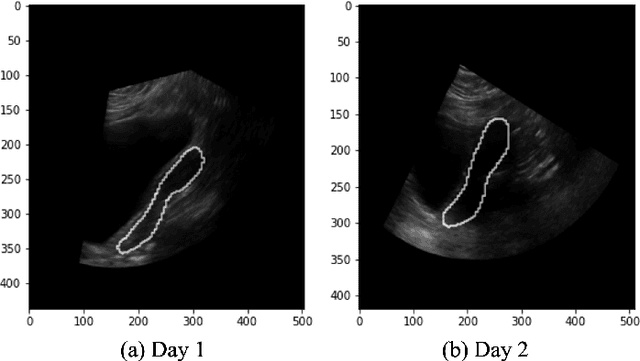
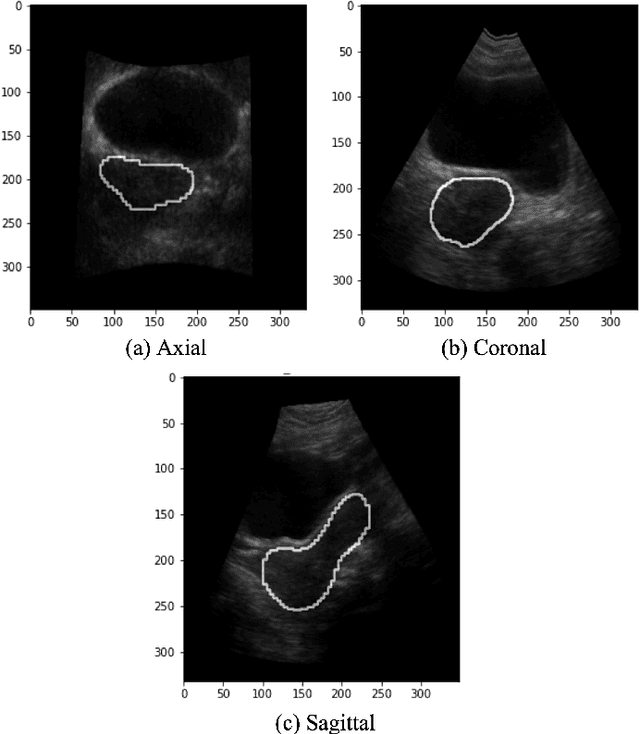
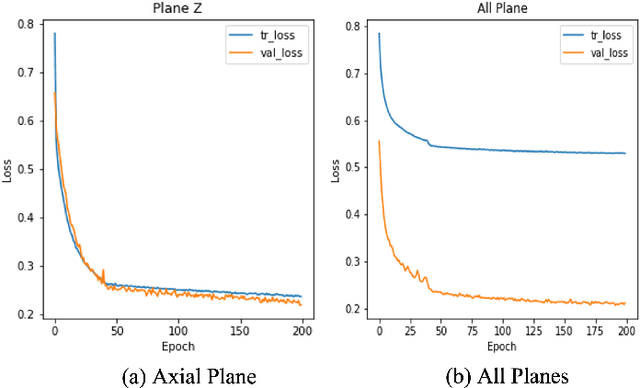
Abstract:On-line segmentation of the uterus can aid effective image-based guidance for precise delivery of dose to the target tissue (the uterocervix) during cervix cancer radiotherapy. 3D ultrasound (US) can be used to image the uterus, however, finding the position of uterine boundary in US images is a challenging task due to large daily positional and shape changes in the uterus, large variation in bladder filling, and the limitations of 3D US images such as low resolution in the elevational direction and imaging aberrations. Previous studies on uterus segmentation mainly focused on developing semi-automatic algorithms where require manual initialization to be done by an expert clinician. Due to limited studies on the automatic 3D uterus segmentation, the aim of the current study was to overcome the need for manual initialization in the semi-automatic algorithms using the recent deep learning-based algorithms. Therefore, we developed 2D UNet-based networks that are trained based on two scenarios. In the first scenario, we trained 3 different networks on each plane (i.e., sagittal, coronal, axial) individually. In the second scenario, our proposed network was trained using all the planes of each 3D volume. Our proposed schematic can overcome the initial manual selection of previous semi-automatic algorithm.
Breast lesion segmentation in ultrasound images with limited annotated data
Jan 21, 2020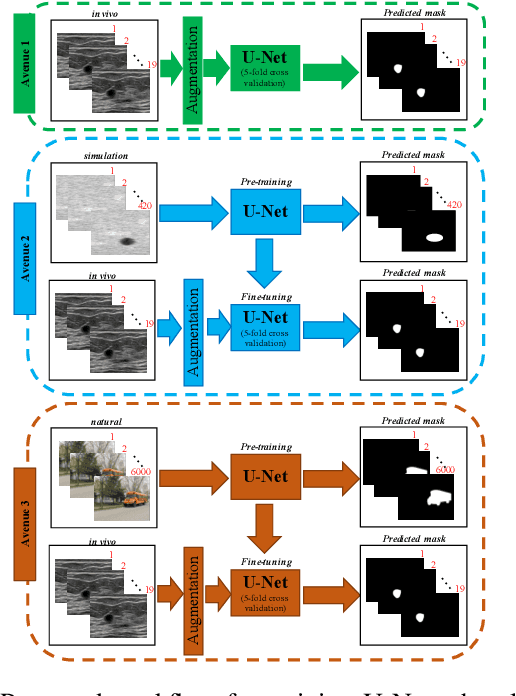
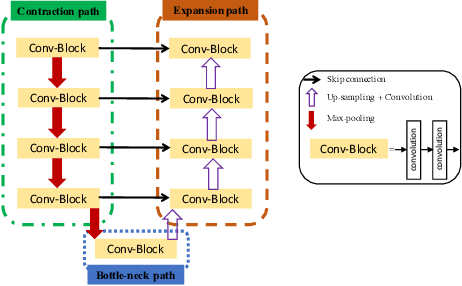
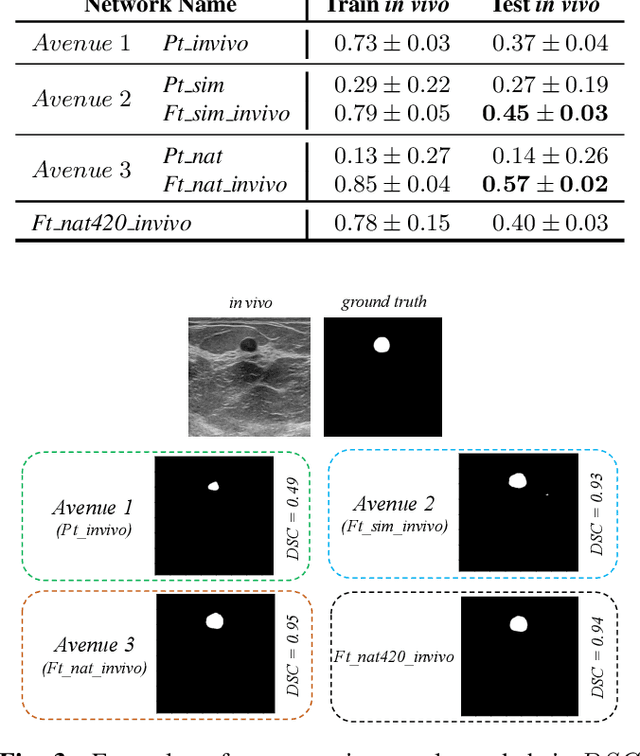
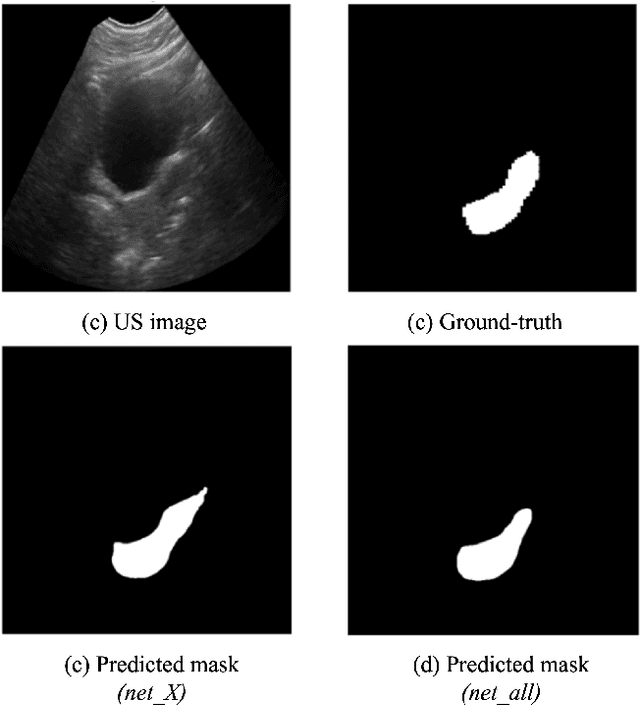
Abstract:Ultrasound (US) is one of the most commonly used imaging modalities in both diagnosis and surgical interventions due to its low-cost, safety, and non-invasive characteristic. US image segmentation is currently a unique challenge because of the presence of speckle noise. As manual segmentation requires considerable efforts and time, the development of automatic segmentation algorithms has attracted researchers attention. Although recent methodologies based on convolutional neural networks have shown promising performances, their success relies on the availability of a large number of training data, which is prohibitively difficult for many applications. Therefore, in this study we propose the use of simulated US images and natural images as auxiliary datasets in order to pre-train our segmentation network, and then to fine-tune with limited in vivo data. We show that with as little as 19 in vivo images, fine-tuning the pre-trained network improves the dice score by 21% compared to training from scratch. We also demonstrate that if the same number of natural and simulation US images is available, pre-training on simulation data is preferable.
Ultrasound segmentation using U-Net: learning from simulated data and testing on real data
Apr 24, 2019



Abstract:Segmentation of ultrasound images is an essential task in both diagnosis and image-guided interventions given the ease-of-use and low cost of this imaging modality. As manual segmentation is tedious and time consuming, a growing body of research has focused on the development of automatic segmentation algorithms. Deep learning algorithms have shown remarkable achievements in this regard; however, they need large training datasets. Unfortunately, preparing large labeled datasets in ultrasound images is prohibitively difficult. Therefore, in this study, we propose the use of simulated ultrasound (US) images for training the U-Net deep learning segmentation architecture and test on tissue-mimicking phantom data collected by an ultrasound machine. We demonstrate that the trained architecture on the simulated data is transferrable to real data, and therefore, simulated data can be considered as an alternative training dataset when real datasets are not available. The second contribution of this paper is that we train our U- Net network on envelope and B-mode images of the simulated dataset, and test the trained network on real envelope and B- mode images of phantom, respectively. We show that test results are superior for the envelope data compared to B-mode image.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge