Ashish Ranjan
Scalable Permutation-Aware Modeling for Temporal Set Prediction
Apr 23, 2025Abstract:Temporal set prediction involves forecasting the elements that will appear in the next set, given a sequence of prior sets, each containing a variable number of elements. Existing methods often rely on intricate architectures with substantial computational overhead, which hampers their scalability. In this work, we introduce a novel and scalable framework that leverages permutation-equivariant and permutation-invariant transformations to efficiently model set dynamics. Our approach significantly reduces both training and inference time while maintaining competitive performance. Extensive experiments on multiple public benchmarks show that our method achieves results on par with or superior to state-of-the-art models across several evaluation metrics. These results underscore the effectiveness of our model in enabling efficient and scalable temporal set prediction.
Pneumonia Detection in Chest X-Rays using Neural Networks
Apr 07, 2022
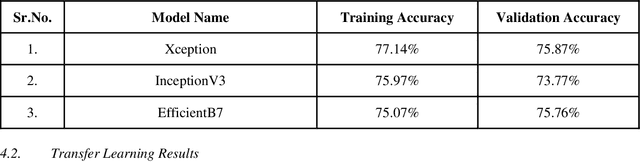
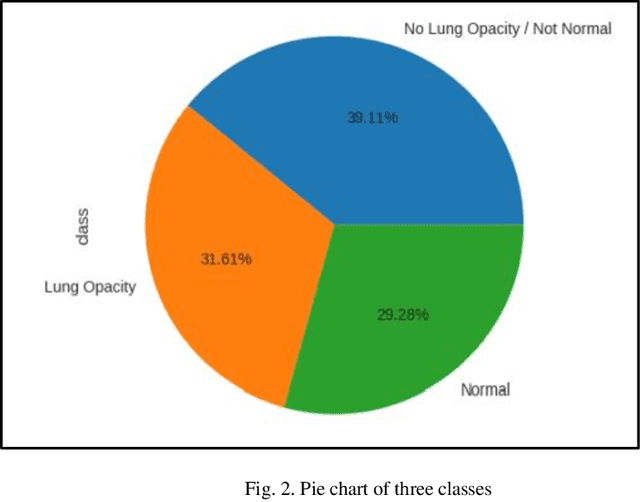
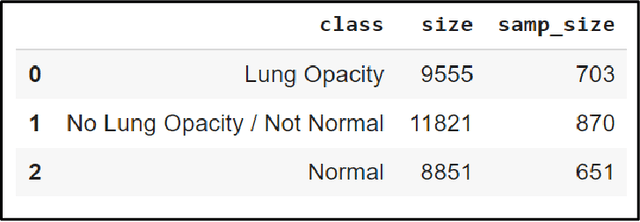
Abstract:With the advancement in AI, deep learning techniques are widely used to design robust classification models in several areas such as medical diagnosis tasks in which it achieves good performance. In this paper, we have proposed the CNN model (Convolutional Neural Network) for the classification of Chest X-ray images for Radiological Society of North America Pneumonia (RSNA) datasets. The study also tries to achieve the same RSNA benchmark results using the limited computational resources by trying out various approaches to the methodologies that have been implemented in recent years. The proposed method is based on a non-complex CNN and the use of transfer learning algorithms like Xception, InceptionV3/V4, EfficientNetB7. Along with this, the study also tries to achieve the same RSNA benchmark results using the limited computational resources by trying out various approaches to the methodologies that have been implemented in recent years. The RSNA benchmark MAP score is 0.25, but using the Mask RCNN model on a stratified sample of 3017 along with image augmentation gave a MAP score of 0.15. Meanwhile, the YoloV3 without any hyperparameter tuning gave the MAP score of 0.32 but still, the loss keeps decreasing. Running the model for a greater number of iterations can give better results.
λ-Scaled-Attention: A Novel Fast Attention Mechanism for Efficient Modeling of Protein Sequences
Jan 09, 2022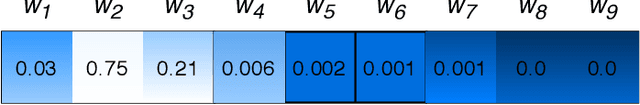
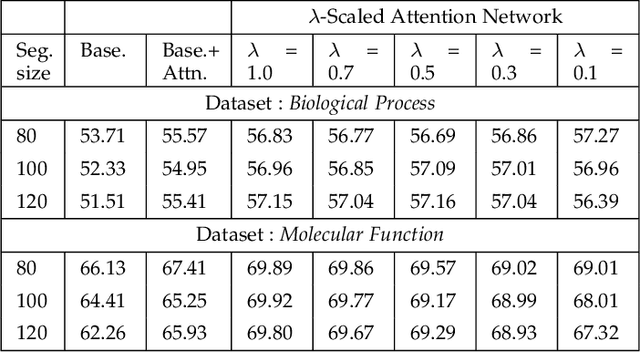
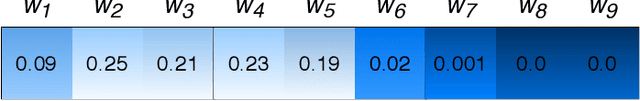
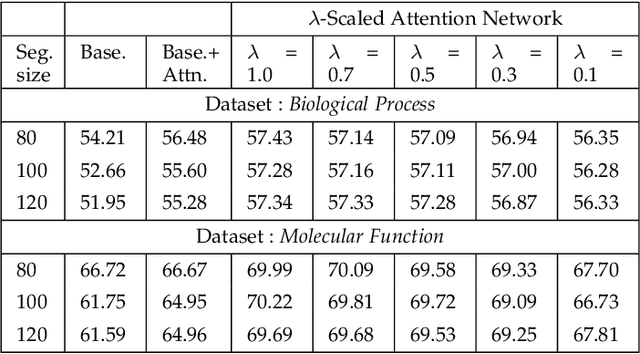
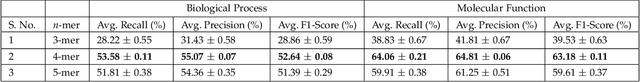
Abstract:Attention-based deep networks have been successfully applied on textual data in the field of NLP. However, their application on protein sequences poses additional challenges due to the weak semantics of the protein words, unlike the plain text words. These unexplored challenges faced by the standard attention technique include (i) vanishing attention score problem and (ii) high variations in the attention distribution. In this regard, we introduce a novel {\lambda}-scaled attention technique for fast and efficient modeling of the protein sequences that addresses both the above problems. This is used to develop the {\lambda}-scaled attention network and is evaluated for the task of protein function prediction implemented at the protein sub-sequence level. Experiments on the datasets for biological process (BP) and molecular function (MF) showed significant improvements in the F1 score values for the proposed {\lambda}-scaled attention technique over its counterpart approach based on the standard attention technique (+2.01% for BP and +4.67% for MF) and state-of-the-art ProtVecGen-Plus approach (+2.61% for BP and +4.20% for MF). Further, fast convergence (converging in half the number of epochs) and efficient learning (in terms of very low difference between the training and validation losses) were also observed during the training process.
Using a Bi-directional LSTM Model with Attention Mechanism trained on MIDI Data for Generating Unique Music
Nov 02, 2020



Abstract:Generating music is an interesting and challenging problem in the field of machine learning. Mimicking human creativity has been popular in recent years, especially in the field of computer vision and image processing. With the advent of GANs, it is possible to generate new similar images, based on trained data. But this cannot be done for music similarly, as music has an extra temporal dimension. So it is necessary to understand how music is represented in digital form. When building models that perform this generative task, the learning and generation part is done in some high-level representation such as MIDI (Musical Instrument Digital Interface) or scores. This paper proposes a bi-directional LSTM (Long short-term memory) model with attention mechanism capable of generating similar type of music based on MIDI data. The music generated by the model follows the theme/style of the music the model is trained on. Also, due to the nature of MIDI, the tempo, instrument, and other parameters can be defined, and changed, post generation.
A Parallel Approach for Real-Time Face Recognition from a Large Database
Nov 01, 2020



Abstract:We present a new facial recognition system, capable of identifying a person, provided their likeness has been previously stored in the system, in real time. The system is based on storing and comparing facial embeddings of the subject, and identifying them later within a live video feed. This system is highly accurate, and is able to tag people with their ID in real time. It is able to do so, even when using a database containing thousands of facial embeddings, by using a parallelized searching technique. This makes the system quite fast and allows it to be highly scalable.
Deep Robust Framework for Protein Function Prediction using Variable-Length Protein Sequences
Nov 04, 2018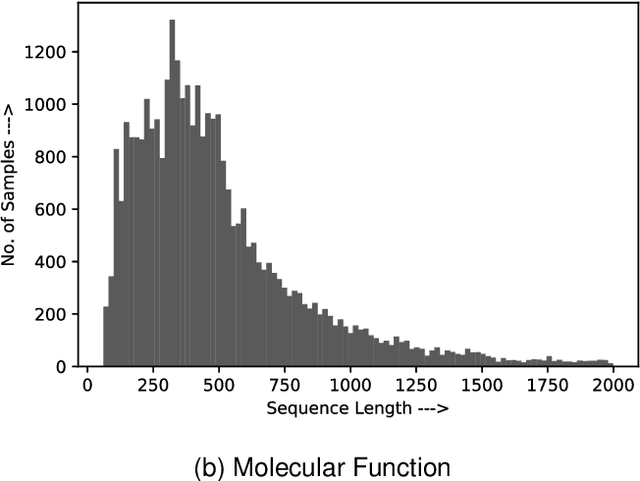
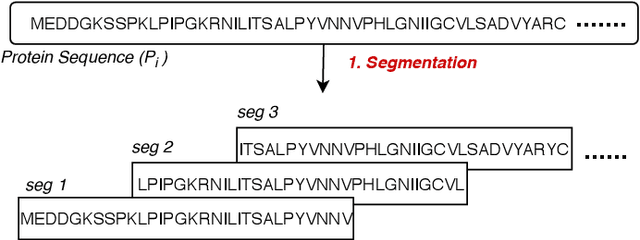
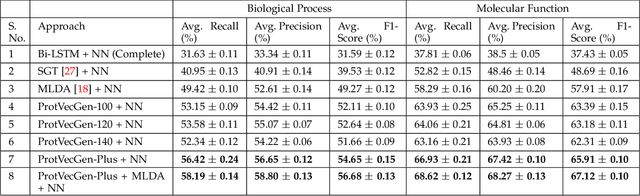
Abstract:Amino acid sequence portrays most intrinsic form of a protein and expresses primary structure of protein. The order of amino acids in a sequence enables a protein to acquire a particular stable conformation that is responsible for the functions of the protein. This relationship between a sequence and its function motivates the need to analyse the sequences for predicting protein functions. Early generation computational methods using BLAST, FASTA, etc. perform function transfer based on sequence similarity with existing databases and are computationally slow. Although machine learning based approaches are fast, they fail to perform well for long protein sequences (i.e., protein sequences with more than 300 amino acid residues). In this paper, we introduce a novel method for construction of two separate feature sets for protein sequences based on analysis of 1) single fixed-sized segments and 2) multi-sized segments, using bi-directional long short-term memory network. Further, model based on proposed feature set is combined with the state of the art Multi-lable Linear Discriminant Analysis (MLDA) features based model to improve the accuracy. Extensive evaluations using separate datasets for biological processes and molecular functions demonstrate promising results for both single-sized and multi-sized segments based feature sets. While former showed an improvement of +3.37% and +5.48%, the latter produces an improvement of +5.38% and +8.00% respectively for two datasets over the state of the art MLDA based classifier. After combining two models, there is a significant improvement of +7.41% and +9.21% respectively for two datasets compared to MLDA based classifier. Specifically, the proposed approach performed well for the long protein sequences and superior overall performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge