λ-Scaled-Attention: A Novel Fast Attention Mechanism for Efficient Modeling of Protein Sequences
Paper and Code
Jan 09, 2022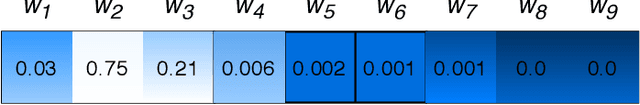
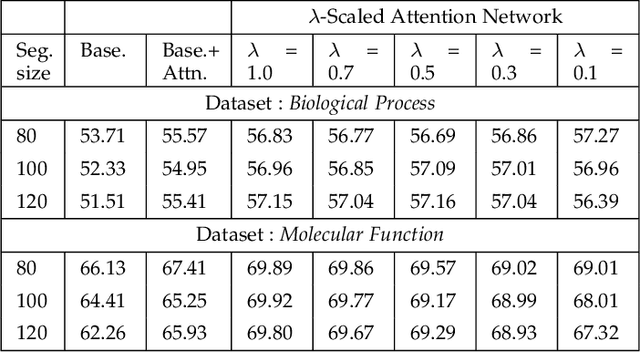
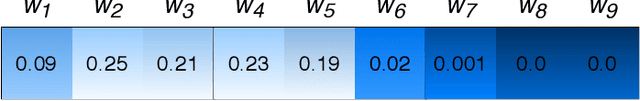
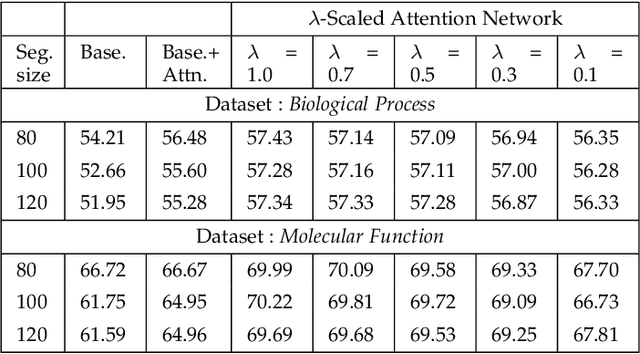
Attention-based deep networks have been successfully applied on textual data in the field of NLP. However, their application on protein sequences poses additional challenges due to the weak semantics of the protein words, unlike the plain text words. These unexplored challenges faced by the standard attention technique include (i) vanishing attention score problem and (ii) high variations in the attention distribution. In this regard, we introduce a novel {\lambda}-scaled attention technique for fast and efficient modeling of the protein sequences that addresses both the above problems. This is used to develop the {\lambda}-scaled attention network and is evaluated for the task of protein function prediction implemented at the protein sub-sequence level. Experiments on the datasets for biological process (BP) and molecular function (MF) showed significant improvements in the F1 score values for the proposed {\lambda}-scaled attention technique over its counterpart approach based on the standard attention technique (+2.01% for BP and +4.67% for MF) and state-of-the-art ProtVecGen-Plus approach (+2.61% for BP and +4.20% for MF). Further, fast convergence (converging in half the number of epochs) and efficient learning (in terms of very low difference between the training and validation losses) were also observed during the training process.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge